Tackling hepatitis B Virus with CRISPR/Cas9: advances, challenges, and delivery strategies
- Review Paper
- Published: 28 August 2024

Cite this article

- Dakshina M. Nair ORCID: orcid.org/0009-0009-0922-0142 1 ,
- Leela Kakithakara Vajravelu 1 ,
- Jayaprakash Thulukanam 1 ,
- Vishnupriya Paneerselvam 1 ,
- Poornima Baskar Vimala 1 &
- Rahul Harikumar Lathakumari 1
121 Accesses
Explore all metrics
Hepatitis B virus (HBV) infection remains a significant global health challenge, with chronic HBV leading to severe liver diseases, including cirrhosis and hepatocellular carcinoma. Current treatments often fail to eradicate the virus, highlighting the need for innovative therapeutic strategies. The CRISPR/Cas9 system has emerged as a dynamic tool for precise genome editing and presents a promising approach to targeting and eliminating HBV infection. This review provides a comprehensive overview of the advances, challenges, and delivery strategies associated with CRISPR/Cas9-based therapies for HBV. We begin by elucidating the mechanism of the CRISPR/Cas9 system and then explore HBV pathogenesis, focusing on the role of covalently closed circular DNA (cccDNA) and integrated HBV DNA in maintaining chronic infection. CRISPR/Cas9 can disrupt these key viral reservoirs, which are critical for persistent HBV replication and associated liver damage. The application of CRISPR/Cas9 in HBV treatment faces significant challenges, such as off-target effects, delivery efficiency, and immune responses. These challenges are addressed by examining current approaches to enhance the specificity, safety, and efficacy of CRISPR/Cas9. A future perspective on the development and clinical translation of CRISPR/Cas9 therapies for HBV is provided, emphasizing the requirement for further research to improve delivery methods and ensure durable safety and effectiveness. This review underscores the transformative potential of CRISPR/Cas9 in combating HBV and sets the stage for future breakthroughs in the field.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Subscribe and save.
- Get 10 units per month
- Download Article/Chapter or eBook
- 1 Unit = 1 Article or 1 Chapter
- Cancel anytime
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions

Similar content being viewed by others

The Role of RNA Interference in Functional Cure Strategies for Chronic Hepatitis B

Recent Advances in Use of Gene Therapy to Treat Hepatitis B Virus Infection
The impact of integrated hepatitis B virus DNA on oncogenesis and antiviral therapy
Data availability.
No datasets were generated or analyzed during the current study.
Fletcher GJ, Eapen CE, Abraham P (2020) Hepatitis B genotyping: the utility for the clinicians. Indian J Gastroenterol 39(4):315–320. https://doi.org/10.1007/s12664-019-00995-y
Article PubMed Google Scholar
Kong H, Ju E, Yi K, Xu W, Lao YH, Cheng D, Zhang Q, Tao Y, Li M, Ding J (2021) Advanced nanotheranostics of CRISPR/Cas for viral hepatitis and hepatocellular carcinoma. Adv Sci (Weinh). 8:e2102051. https://doi.org/10.1002/advs.202102051
Article CAS PubMed Google Scholar
Simmonds P (2001) The origin and evolution of hepatitis viruses in humans. J Gen Virol 82(Pt 4):693–712. https://doi.org/10.1099/0022-1317-82-4-693
Biswas A, Banerjee A, Chandra PK, Datta S, Panigrahi R, Dutta D, De BK, Pal M, Guha SK, Chakrabarti S, Chakravarty R (2011) Variations in the functional domain of basal core promoter of hepatitis B virus among Eastern Indian patients with prevalence of genotypes A, C, and D among the same ethnic population. J Med Virol 83(2):253–260. https://doi.org/10.1002/jmv.21979
Piermatteo L, D’Anna S, Bertoli A, Bellocchi M, Carioti L, Fabeni L, Alkhatib M, Frazia S, Lichtner M, Mastroianni C, Sanctis G, Marignani M, Pasquazzi C, Iapadre N, Parruti G, Cappiello G, Vecchiet J, Malagnino V, Grelli S, Ceccherini-Silbertein F, Andreoni M, Sarmati L, Svicher V, Salpini R (2023) Unexpected rise in the circulation of complex HBV variants enriched of HBsAg vaccine-escape mutations in HBV genotype-D: potential impact on HBsAg detection/quantification and vaccination strategies. Emerg Microbes Infect 12(1):2219347. https://doi.org/10.1080/22221751.2023.2219347
Article CAS PubMed PubMed Central Google Scholar
Block TM, Rawat S, Brosgart CL (2015) Chronic hepatitis B: a wave of new therapies on the horizon. Antiviral Res. 121:69–81. https://doi.org/10.1016/j.antiviral.2015.06.014
Zeisel MB, Lucifora J, Mason WS, Sureau C, Beck J, Levrero M, Kann M, Knolle PA, Benkirane M, Durantel D, Michel ML, Autran B, Cosset FL, Strick-Marchand H, Trépo C, Kao JH, Carrat F, Lacombe K, Schinazi RF, Barré-Sinoussi F, Delfraissy JF, Zoulim F (2015) Towards an HBV cure: state-of-the-art and unresolved questions–report of the ANRS workshop on HBV cure. Gut 64(8):1314–1326. https://doi.org/10.1136/gutjnl-2014-308943
Lai CL, Shouval D, Lok AS, Chang TT, Cheinquer H, Goodman Z, DeHertogh D, Wilber R, Zink RC, Cross A, Colonno R, Fernandes L, BEHoLD AI463027 study group (2006) Entecavir versus lamivudine for patients with HBeAg-negative chronic hepatitis B. N Engl J Med. 354(10):1011–20. https://doi.org/10.1056/NEJMoa051287
Kar P, Goswami B, Mahanta J, Bhimo T, Das AK, Deka M, Lynrah KG, Kotwal MR, Bhaumik P, Jini M, Karna R, Karra VK, Kaur H (2022) Epidemiology, Genotyping, mutational and phylogenetic analysis of hepatitis B virus infection in North-East India. J Clin Exp Hepatol. 12(1):43–51. https://doi.org/10.1016/j.jceh.2021.04.002
Siederdissen HZC, Cornberg M (2016) Management of HBV and HBV/HDV-associated liver cirrhosis. Visc Med. 32(2):86–94. https://doi.org/10.1159/000445518
Article Google Scholar
Wursthorn K, Lutgehetmann M, Dandri M, Volz T, Buggisch P, Zollner B, Longerich T, Schirmacher P, Metzler F, Zankel M, Fischer C, Currie G, Brosgart C, Petersen J (2006) Peginterferon alpha-2b plus adefovir induce strong cccDNA decline and HBsAg reduction in patients with chronic hepatitis B. Hepatology 44(3):675–684. https://doi.org/10.1002/hep.21282
Lin G, Zhang K, Li J (2015) Application of CRISPR/Cas9 technology to HBV. Int J Mol Sci 16(11):26077–26086. https://doi.org/10.3390/ijms161125950.
Liu H, Zakrzewicz D, Nosol K, Irobalieva RN, Mukherjee S, Bang-Sørensen R, Goldmann N, Kunz S, Rossi L, Kossiakoff AA, Urban S, Glebe D, Geyer J, Locher KP (2024) Structure of antiviral drug bulevirtide bound to hepatitis B and D virus receptor protein NTCP. Nat Commun 15(1):2476. https://doi.org/10.1038/s41467-024-46706-w
Nassal M (2015) HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64(12):1972–1984. https://doi.org/10.1136/gutjnl-2015-309809
Seeger C, Sohn JA (2014) Targeting Hepatitis B Virus With CRISPR/Cas9. Mol Ther Nucleic Acids 3(12):e216. https://doi.org/10.1038/mtna.2014.68
Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A (1987) Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 169(12):5429–5433. https://doi.org/10.1128/jb.169.12.5429-5433.1987
Mojica FJ, Juez G, Rodríguez-Valera F (1993) Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol 9(3):613–621. https://doi.org/10.1111/j.1365-2958.1993.tb01721.x
Asmamaw M, Zawdie B (2021) Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biologics 21(15):353–361. https://doi.org/10.2147/BTT.S326422
Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV (2002) A DNA repair system specific for thermophilic archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res 30(2):482–496. https://doi.org/10.1093/nar/30.2.482
Jansen R, Embden JD, Gaastra W, Schouls LM (2002) Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43(6):1565–1575. https://doi.org/10.1046/j.1365-2958.2002.02839.x
Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV (2006) A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 16(1):7. https://doi.org/10.1186/1745-6150-1-7
Article CAS Google Scholar
Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E (2005) Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60(2):174–182. https://doi.org/10.1007/s00239-004-0046-3
Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321(5891):960–964. https://doi.org/10.1126/science.1159689
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471(7340):602–607. https://doi.org/10.1038/nature09886
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. https://doi.org/10.1126/science.1225829
Article PubMed PubMed Central Google Scholar
Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Reading) 155(Pt 3):733–740. https://doi.org/10.1099/mic.0.023960-0
Mussolino C, Cathomen T (2013) RNA guides genome engineering. Nat Biotechnol 31(3):208–209. https://doi.org/10.1038/nbt.2527
Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA (2013) Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 10:741–3. https://doi.org/10.1038/nmeth.2532
Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK (2013) Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 31:227–9. https://doi.org/10.1038/nbt.2501
Port F, Chen HM, Lee T, Bullock SL (2014) Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A. 111:E2967-76. https://doi.org/10.1073/pnas.1405500111
Tu T, Douglas MW (2020) Hepatitis B virus infection: from diagnostics to treatments. Viruses 12(12):1366. https://doi.org/10.3390/v12121366
Iannacone M, Guidotti LG (2022) Immunobiology and pathogenesis of hepatitis B virus infection. Nat Rev Immunol 22(1):19–32. https://doi.org/10.1038/s41577-021-00549-4
Wang J, Xu ZW, Liu S, Zhang RY, Ding SL, Xie XM, Long L, Chen XM, Zhuang H, Lu FM (2015) Dual gRNAs guided CRISPR/Cas9 system inhibits hepatitis B virus replication. World J Gastroenterol 21(32):9554–9565. https://doi.org/10.3748/wjg.v21.i32.9554
Dong C, Qu L, Wang H, Wei L, Dong Y, Xiong S (2015) Targeting hepatitis B virus cccDNA by CRISPR/Cas9 nuclease efficiently inhibits viral replication. Antiviral Res 118:110–117. https://doi.org/10.1016/j.antiviral.2015.03.015
Seeger C, Mason WS (2015) Molecular biology of hepatitis B virus infection. Virology. https://doi.org/10.1016/j.virol.2015.02.031
Zhou Z, Li C, Tan Z, Sun G, Peng B, Ren T, He J, Wang Y, Sun Y, Wang F, Li W (2023) A spatiotemporally controlled recombinant cccDNA mouse model for studying HBV and developing drugs against the virus. Antiviral Res 216:105642. https://doi.org/10.1016/j.antiviral.2023.105642
Ramanan V, Shlomai A, Cox DB, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN (2015) CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep 2(5):10833. https://doi.org/10.1038/srep10833
Yang YC, Yang HC (2021) Recent progress and future prospective in HBV cure by CRISPR/Cas. Viruses 14(1):4. https://doi.org/10.3390/v14010004
Doudna JA, Charpentier E, Genome editing (2014) The new frontier of genome engineering with CRISPR-Cas9. Science. https://doi.org/10.1126/science.1258096
Kostyushev D, Brezgin S, Kostyusheva A, Zarifyan D, Goptar I, Chulanov V (2019) Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus. Cell Mol Life Sci 76(9):1779–1794. https://doi.org/10.1007/s00018-019-03021-8
Liu X, Hao R, Chen S, Guo D, Chen Y (2015) Inhibition of hepatitis B virus by the CRISPR/Cas9 system via targeting the conserved regions of the viral genome. J Gen Virol 96(8):2252–2261. https://doi.org/10.1099/vir.0.000159
Martinez MG, Smekalova E, Combe E, Gregoire F, Zoulim F, Testoni B (2022) Gene editing technologies to target HBV cccDNA. Viruses 14(12):2654. https://doi.org/10.3390/v14122654
Mehta A, Merkel OM (2020) Immunogenicity of Cas9 protein. J Pharm Sci. 109(1):62–67. https://doi.org/10.1016/j.xphs.2019.10.003
Kosicki M, Tomberg K, Bradley A (2018) Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat Biotechnol. 36(8):765–771. https://doi.org/10.1038/nbt.4192
Li H, Sheng C, Wang S, Yang L, Liang Y, Huang Y, Liu H, Li P, Yang C, Yang X, Jia L, Xie J, Wang L, Hao R, Du X, Xu D, Zhou J, Li M, Sun Y, Tong Y, Li Q, Qiu S, Song H (2017) Removal of integrated hepatitis B Virus DNA using CRISPR-Cas9. Front Cell Infect Microbiol 22(7):91. https://doi.org/10.3389/fcimb.2017.00091
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 556(7699):57–63. https://doi.org/10.1038/nature26155
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK (2016) High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529(7587):490–5. https://doi.org/10.1038/nature16526
Martinez MG, Combe E, Inchauspe A, Mangeot PE, Delberghe E, Chapus F, Neveu G, Alam A, Carter K, Testoni B, Zoulim F (2022) CRISPR-Cas9 targeting of hepatitis B virus covalently closed circular DNA generates transcriptionally active episomal variants. mBio. 13:e0288821. https://doi.org/10.1128/mbio.02888-21
Hu P, Li Y, Zhang W, Liu R, Peng L, Xu R, Cai J, Yuan H, Feng T, Tian A, Yue M, Li J, Li W, Zhu C (2023) The spliceosome factor EFTUD2 promotes IFN Anti-HBV effect through mRNA splicing. Mediators Inflamm 23(2023):2546278. https://doi.org/10.1155/2023/2546278
Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O (2018) Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 361(6408):1259–1262. https://doi.org/10.1126/science.aas9129
Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F (2015) In vivo genome editing using staphylococcus aureus Cas9. Nature. 520(7546):186–91. https://doi.org/10.1038/nature14299
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 163(3):759–71. https://doi.org/10.1016/j.cell.2015.09.038
Kim E, Koo T, Park SW, Kim D, Kim K, Cho HY, Song DW, Lee KJ, Jung MH, Kim S, Kim JH, Kim JH, Kim JS (2017) In vivo genome editing with a small Cas9 orthologue derived from campylobacter jejuni. Nat Commun 21(8):14500. https://doi.org/10.1038/ncomms14500
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F (2016) Rationally engineered Cas9 nucleases with improved specificity. Science. 351(6268):84–8. https://doi.org/10.1126/science.aad5227
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5):1173–1183
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 533(7603):420–4. https://doi.org/10.1038/nature17946
Hu B, Zou Y, Zhang L, Tang J, Niedermann G, Firat E, Huang X, Zhu X (2019) Nucleofection with plasmid DNA for CRISPR/Cas9-mediated inactivation of programmed cell death protein 1 in CD133-specific CAR T cells. Hum Gene Ther 30(4):446–458. https://doi.org/10.1089/hum.2017.
Hendriks WT, Jiang X, Daheron L, Cowan CA (2015) TALEN- and CRISPR/Cas9-mediated gene editing in human pluripotent stem cells using lipid-based transfection. Curr Protoc Stem Cell Biol. https://doi.org/10.1002/9780470151808.sc05b03s34
Horii T, Arai Y, Yamazaki M, Morita S, Kimura M, Itoh M, Abe Y, Hatada I (2014) Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep 28(4):4513. https://doi.org/10.1038/srep04513
Carey K, Ryu J, Uh K, Lengi AJ, Clark-Deener S, Corl BA, Lee K (2019) Frequency of off-targeting in genome edited pigs produced via direct injection of the CRISPR/Cas9 system into developing embryos. BMC Biotechnol 19(1):25. https://doi.org/10.1186/s12896-019-0517-7
McAndrews KM, Xiao F, Chronopoulos A, LeBleu VS, Kugeratski FG, Kalluri R (2021) Exosome-mediated delivery of CRISPR/Cas9 for targeting of oncogenic Kras G12D in pancreatic cancer. Life Sci Alliance 4(9):e202000875. https://doi.org/10.26508/lsa.202000875
Alallam B, Altahhan S, Taher M, Mohd Nasir MH, Doolaanea AA (2020) Electrosprayed alginate nanoparticles as CRISPR plasmid DNA delivery carrier: preparation, optimization, and characterization. Pharmaceuticals (Basel) 13(8):158. https://doi.org/10.3390/ph13080158
Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Hüser N, Durantel D, Liang TJ, Münk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U (2014) Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 343(6176):1221–8. https://doi.org/10.1126/science.1243462
Xia Y, Stadler D, Lucifora J, Reisinger F, Webb D, Hösel M, Michler T, Wisskirchen K, Cheng X, Zhang K, Chou WM, Wettengel JM, Malo A, Bohne F, Hoffmann D, Eyer F, Thimme R, Falk CS, Thasler WE, Heikenwalder M, Protzer U (2016) Interferon-γ and tumor necrosis factor-α produced by T cells reduce the HBV persistence form, cccDNA. Without Cytol Gastroenterol 150(1):194–205. https://doi.org/10.1053/j.gastro.2015.09.026
Kruse RL, Shum T, Tashiro H, Barzi M, Yi Z, Whitten-Bauer C, Legras X, Bissig-Choisat B, Garaigorta U, Gottschalk S, Bissig KD (2018) HBsAg-redirected T cells exhibit antiviral activity in HBV-infected human liver chimeric mice. Cytotherapy. 20(5):697–705. https://doi.org/10.1016/j.jcyt.2018.02.002
Li X, Zhao J, Yuan Q, Xia N (2017) Detection of HBV covalently closed circular DNA. Viruses 9(6):139. https://doi.org/10.3390/v9060139
Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, Wu FY, Kao JH, Chen DS, Chen PJ (2014) The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates In Vivo. Mol Ther Nucleic Acids 3(8):e186. https://doi.org/10.1038/mtna.2014.38.PMID:25137139;PMCID:PMC4221598
Kayesh MEH, Amako Y, Hashem MA, Murakami S, Ogawa S, Yamamoto N, Hifumi T, Miyoshi N, Sugiyama M, Tanaka Y, Mizokami M, Kohara M, Tsukiyama-Kohara K (2020) Development of an in vivo delivery system for CRISPR/Cas9-mediated targeting of hepatitis B virus cccDNA. Virus Res 290:198191. https://doi.org/10.1016/j.virusres.2020.198191
Sakuma T, Masaki K, Abe-Chayama H, Mochida K, Yamamoto T, Chayama K (2016) Highly multiplexed CRISPR-Cas9-nuclease and Cas9-nickase vectors for inactivation of hepatitis B virus. Genes Cells 21(11):1253–1262. https://doi.org/10.1111/gtc.12437
Kurihara T, Fukuhara T, Ono C, Yamamoto S, Uemura K, Okamoto T, Sugiyama M, Motooka D, Nakamura S, Ikawa M, Mizokami M, Maehara Y, Matsuura Y (2017) Suppression of HBV replication by the expression of nickase- and nuclease dead-Cas9. Sci Rep 7(1):6122. https://doi.org/10.1038/s41598-017-05905-w
Liu Y, Zhao M, Gong M, Xu Y, Xie C, Deng H, Li X, Wu H, Wang Z (2018) Inhibition of hepatitis B virus replication via HBV DNA cleavage by Cas9 from staphylococcus aureus. Antiviral Res 152:58–67. https://doi.org/10.1016/j.antiviral.2018.02.011
Karimova M, Beschorner N, Dammermann W, Chemnitz J, Indenbirken D, Bockmann JH, Grundhoff A, Lüth S, Buchholz F, Schulze zur Wiesch J, Hauber J (2015) CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Sci Rep 5:13734. https://doi.org/10.1038/srep13734
Stone D, Long KR, Loprieno MA, De Silva Feelixge HS, Kenkel EJ, Liley RM, Rapp S, Roychoudhury P, Nguyen T, Stensland L, Colón-Thillet R, Klouser LM, Weber ND, Le C, Wagoner J, Goecker EA, Li AZ, Eichholz K, Corey L, Tyrrell DL, Greninger AL, Huang ML, Polyak SJ, Aubert M, Sagartz JE, Jerome KR (2020) CRISPR-Cas9 gene editing of hepatitis B virus in chronically infected humanized mice. Mol Ther Methods Clin Dev 26(20):258–275. https://doi.org/10.1016/j.omtm.2020.11.014
Kostyusheva AP, Brezgin SA, Ponomareva NI, Goptar IA, Nikiforova AV, Gegechkori VI, Poluektova VB, Turkadze KA, Sudina AE, Chulanov VP, Kostyushev DS (2022) Antiviral activity of CRISPR/Cas9 ribonucleoprotein complexes on a hepatitis B virus model In Vivo. Mol Biol (Mosk). 56(6):884
Yang YC, Chen YH, Kao JH, Ching C, Liu IJ, Wang CC, Tsai CH, Wu FY, Liu CJ, Chen PJ, Chen DS, Yang HC (2020) Permanent inactivation of HBV Genomes by CRISPR/Cas9-mediated non-cleavage base editing. Mol Ther Nucleic Acids. 20:480–490. https://doi.org/10.1016/j.omtn.2020.03.005
Zhu W, Xie K, Xu Y, Wang L, Chen K, Zhang L, Fang J (2016) CRISPR/Cas9 produces anti-hepatitis B virus effect in hepatoma cells and transgenic mouse. Virus Res 2(217):125–132. https://doi.org/10.1016/j.virusres.2016.04.003
Download references
Author information
Authors and affiliations.
Department of Microbiology, SRM Medical College Hospital and Research Centre, SRM Institute of Science and Technology, Kattankulathur, Chengalpattu, Tamil Nadu, India
Dakshina M. Nair, Leela Kakithakara Vajravelu, Jayaprakash Thulukanam, Vishnupriya Paneerselvam, Poornima Baskar Vimala & Rahul Harikumar Lathakumari
You can also search for this author in PubMed Google Scholar
Contributions
Dakshina M Nair: Writing—review and editing, Writing—original draft, Visualization, Validation, Resources, Methodology, Data curation, Conceptualization. Leela Kakithakara Vajravelu: Formal analysis, Conceptualization, Supervision. Jayaprakash Thulukanam: Visualization, Validation, Supervision. Vishnupriya Paneerselvam & Poornima Baskar Vimala: Visualization and Validation. Rahul Harikumar Lathakumari:: Formal analysis.
Corresponding author
Correspondence to Dakshina M. Nair .
Ethics declarations
Competing interests.
The authors declare that they do not have any known competing financial interests or personal relationships that could have appeared to influence the findings reported in this paper.
Additional information
Edited by Juergen Richt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Nair, D.M., Vajravelu, L.K., Thulukanam, J. et al. Tackling hepatitis B Virus with CRISPR/Cas9: advances, challenges, and delivery strategies. Virus Genes (2024). https://doi.org/10.1007/s11262-024-02105-3
Download citation
Received : 03 August 2024
Accepted : 22 August 2024
Published : 28 August 2024
DOI : https://doi.org/10.1007/s11262-024-02105-3
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Hepatitis B
- CRISPR/Cas9
- Chronic infection
- Delivery strategies
- Find a journal
- Publish with us
- Track your research
Loading metrics
Open Access
Peer-reviewed
Research Article
A systematic review of Hepatitis B virus (HBV) prevalence and genotypes in Kenya: Data to inform clinical care and health policy
Roles Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Writing – original draft
Affiliations Nuffield Department of Medicine, Medawar Building for Pathogen Research, University of Oxford, Oxford, United Kingdom, Department of Infectious Diseases and Microbiology, John Radcliffe Hospital, Headley Way, Oxford, United Kingdom
Roles Formal analysis, Methodology, Writing – review & editing
Affiliation Nuffield Department of Medicine, Medawar Building for Pathogen Research, University of Oxford, Oxford, United Kingdom
Roles Investigation, Writing – review & editing
Affiliation CA Medlynks Clinic and Laboratory, Nairobi, and Fountain Projects and Research Office, Fountain Health Care Hospital, Eldoret, Kenya
Affiliations KEMRI-Wellcome Trust Research Programme, Kilifi, Kenya, Department of Biochemistry and Biotechnology, Pwani University, Kilifi, Kenya
Roles Formal analysis, Methodology, Supervision, Writing – review & editing
Contributed equally to this work with: Philippa C. Matthews, Anthony O. Etyang
Roles Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Writing – review & editing
* E-mail: [email protected]
Affiliations Nuffield Department of Medicine, Medawar Building for Pathogen Research, University of Oxford, Oxford, United Kingdom, The Francis Crick Institute, London, United Kingdom, Division of Infection and Immunity, University College London, London, London, United Kingdom, Department of Infectious Diseases, University College London Hospital, London, London, United Kingdom
Roles Conceptualization, Investigation, Supervision, Writing – review & editing
Affiliation KEMRI-Wellcome Trust Research Programme, Kilifi, Kenya
- Louise O. Downs,
- Cori Campbell,
- Paul Yonga,
- George Githinji,
- M. Azim Ansari,
- Philippa C. Matthews,
- Anthony O. Etyang

- Published: January 31, 2023
- https://doi.org/10.1371/journal.pgph.0001165
- See the preprint
- Peer Review
- Reader Comments
The aim of this systematic review and meta-analysis is to evaluate available prevalence and viral sequencing data representing chronic hepatitis B (CHB) infection in Kenya. More than 20% of the global disease burden from CHB is in Africa, however there is minimal high quality seroprevalence data from individual countries and little viral sequencing data available to represent the continent. We undertook a systematic review of the prevalence and genetic data available for hepatitis B virus (HBV) in Kenya using the Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) 2020 checklist. We identified 23 studies reporting HBV prevalence and 8 studies that included HBV genetic data published in English between January 2000 and December 2021. We assessed study quality using the Joanna Briggs Institute critical appraisal checklist. Due to study heterogeneity, we divided the studies to represent low, moderate, high and very high-risk for HBV infection, identifying 8, 7, 5 and 3 studies in these groups, respectively. We calculated pooled HBV prevalence within each group and evaluated available sequencing data. Pooled HBV prevalence was 3.4% (95% CI 2.7–4.2%), 6.1% (95% CI 5.1–7.4%), 6.2% (95% CI 4.64–8.2) and 29.2% (95% CI 12.2–55.1), respectively. Study quality was overall low; only three studies detailed sample size calculation and 17/23 studies were cross sectional. Eight studies included genetic information on HBV, with two undertaking whole genome sequencing. Genotype A accounted for 92% of infections. Other genotypes included genotype D (6%), D/E recombinants (1%) or mixed populations (1%). Drug resistance mutations were reported by two studies. There is an urgent need for more high quality seroprevalence and genetic data to represent HBV in Kenya to underpin improved HBV screening, treatment and prevention in order to support progress towards elimination targets.
Citation: Downs LO, Campbell C, Yonga P, Githinji G, Ansari MA, Matthews PC, et al. (2023) A systematic review of Hepatitis B virus (HBV) prevalence and genotypes in Kenya: Data to inform clinical care and health policy. PLOS Glob Public Health 3(1): e0001165. https://doi.org/10.1371/journal.pgph.0001165
Editor: Abraham D. Flaxman, University of Washington, UNITED STATES
Received: May 31, 2022; Accepted: November 28, 2022; Published: January 31, 2023
Copyright: © 2023 Downs et al. This is an open access article distributed under the terms of the Creative Commons Attribution License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability: All the data pertinent to the submission are included in the paper and its citations.
Funding: LD is funded by a Wellcome Clinician PhD fellowship (Grant number BST00070). CC is funded by GlaxoSmithKline (GSK) and the University of Oxford Nuffield Department of Medicine. PCM is funded by Wellcome (ref 110110Z/15/Z), UCL/UCLH NIHR Biomedical Research Centre (BRC) and core funding from the Francis Crick Institute. MAA is supported by a Sir Henry Dale Fellowship jointly funded by the Royal Society and Wellcome (ref 220171/Z/20/Z). For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Competing interests: I have read the journal’s policy and the authors of this manuscript have the following competing interests: CC is partially funded by GlaxoSmithKline. There are no patents, products in development or marketed products associated with this research to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Introduction
Chronic hepatitis B (CHB) accounts for an estimated 90,000 deaths annually across West, East and Southern Africa, where most countries are of medium to high prevalence for CHB (prevalence ≥4%), accounting for around 20% of the worldwide burden of infection [ 1 ]. The World Health Organisation’s (WHO) point prevalence estimate of CHB for Africa is 6.1% (95% CI 4.6–8.5%), but this varies substantially between settings, and high-quality data for individual countries are scarce [ 1 ]. CHB meets many of the WHO criteria for a neglected tropical disease, including disproportionately affecting populations living in poverty, being associated with significant stigma and discrimination, and poor investment in clinical infrastructure and research [ 2 ]. Fewer than 10% of people have access to testing and treatment, leading to delayed diagnosis, with associated risks of advanced liver disease including hepatocellular carcinoma (HCC) [ 1 ].
The Global Health Sector Strategy (GHSS) for viral hepatitis aims to eliminate HBV as a public health threat by 2030 by reducing the incidence of new chronic infections by 90% and reducing mortality by 65% from the 2015 baseline to achieve the 2030 WHO Sustainable Development Goals [ 3 ]. These are ambitious targets, and current estimates indicate they will not be attained in most settings until beyond 2050 [ 4 ]. Detailed seroprevalence data are lacking, but are urgently needed to target testing, treatment, and prevention interventions to the highest risk groups, to allocate resources, and to inform policy.
In Kenya, there is limited information regarding HBV prevalence. Most studies focus on specific groups such as blood donors and those living with HIV, which may not be representative of the general population [ 5 – 7 ]. Other studies have stringent inclusion criteria, meaning important demographic subgroups remain uncharacterised [ 8 ]. HBV testing is not done routinely in Kenya, even in antenatal populations.
Triple HBV vaccine from the age of 6 weeks onwards is recommended by the Kenyan Ministry of Health as a component of the multivalent vaccines rolled out by GAVI within the WHO Expanded Programme for Immunization (EPI). Hep B birth-dose (BD) vaccine for all babies within 24 hours of birth is recommended by the WHO, but has not been adopted by many countries–including Kenya–due to economic and logistical challenges [ 9 ]. However, more data are needed to underpin evidence-based policy in this domain, and there is increasing focus on PMTCT as part of ‘triple elimination’ strategies for HBV/HIV/Syphilis [ 10 ].
HBV is divided into 9 genotypes (A-I) with a 10 th putative genotype J [ 11 , 12 ]; these tend to have distinct geographical locations and have been linked to different outcomes. Genotype A predominates in many African countries and has been associated with horizontal transmission, chronicity, early HBeAg seroconversion [ 13 ], cirrhosis and HCC development [ 14 ]. Genotype also affects response to treatment (including drug resistance), and thus may influence clinical recommendations [ 13 – 15 ], though is not yet widely undertaken in clinical practice in most settings. Most studies of the impact of HBV genotype have been in Asia and Europe. There is a paucity of data on circulating genotypes and subgenotypes in Africa, including Kenya. Whole genome sequencing (WGS) of HBV in Kenya could provide information on transmission networks, disease and treatment outcomes, drug resistance and vaccine escape.
We here assimilate data to describe the seroprevalence and molecular characteristics of HBV infection in Kenya to underpin an evidence-base for local strategies for intervention, and highlight knowledge gaps to inform research. High resolution local data will be essential for development of local clinical care pathways and public health policy, to underpin progress towards the 2030 elimination targets.
Ethics statement
No ethical approval was required for this study.
Search strategy
We set out to review literature on prevalence and genetic characteristics of HBV infection in Kenya, using the Preferred Reporting Items for Systematic Review and Meta-analysis (PRISMA) 2020 statement checklist ( S1 PRISMA Checklist ). We searched the online databases PubMed, Embase, African Journals Online (AJOL) and Scopus on 6 th December 2021 using the terms in Table 1 . We included studies published in English, from 2000 to December 2021 (from 2003 for AJOL) that investigated prevalence, genotype and sequencing of HBV infection in Kenya. We only included data for adults from studies for which the full text was available. There was no minimum number of participants for studies included. We initially screened using a thorough review of the title and abstract, and subsequently reviewed the full manuscripts of eligible articles. Articles that did not meet the inclusion criteria were excluded. Any uncertainty regarding the inclusion of papers was discussed with another reviewer and a consensus obtained.
- PPT PowerPoint slide
- PNG larger image
- TIFF original image
https://doi.org/10.1371/journal.pgph.0001165.t001
From each study, we extracted:
- Total number of individuals tested for HBV.
- Number of individuals found to be infected with HBV (either HBsAg positive or HBV DNA positive)
- Study location (city or geographical region)
- Participant selection criteria
- Laboratory methods for confirmation of HBV infection
- Whether any viral sequencing was undertaken, methods used and results (including genotype, presence of vaccine escape and drug resistance mutations).
Study heterogeneity and HBV risk groups
On the grounds of significant heterogeneity in the populations represented, we divided studies a priori into four groups representing populations with differing risks of testing positive for HBV infection. The low-risk group included studies likely to be most representative of the general population (antenatal women, healthcare workers, blood donors and the national survey). The moderate risk group consisted of studies containing populations living with HIV. High-risk groups were defined as people with risk factors for acquisition of blood-borne virus infection, including people who inject drugs, men who have sex with men (MSM) and sex workers. Those presenting to hospital with hepatitis or jaundice were defined as a very high-risk group, as HBV infection is enriched in populations presenting with established liver disease, particularly if the background population has medium or high HBV prevalence. This risk stratification system is a pragmatic approach to a highly heterogenous literature and we have used these risk groups for ease of reference throughout this review.
Quality assessment of studies
A thorough assessment of the study quality was done using the PRISMA guidelines [ 16 ] and Joanna Briggs Institute critical appraisal checklist for prevalence studies ( S1 Table ) [ 17 ]. Any dispute surrounding study quality was discussed with another reviewer and a consensus reached.
Identifying and analysing full-length HBV sequences from Kenya
We downloaded all full genome HBV sequences from Kenya in GenBank on 1-December-2021 to assimilate a reference set of all whole genome sequences representing Kenya. Sequences were aligned with available HBV reference sequences for each genotype (11) using MAFFT [ 18 ]. A maximum likelihood phylogenetic tree with bootstrap replicates of 1000 was created using NGPhylogeny.fr [ 19 ].
Statistical analysis
Occult HBV infection
Occult HBV infection (OBI) is defined as detectable HBV DNA in the absence of HBsAg. Where studies reported both HBsAg positivity rates and OBI rates in those who were HBsAg negative, only prevalence data based on HBsAg positivity was included in the meta-analysis, in order to ensure datasets were comparable between studies.
(i) Identification of studies
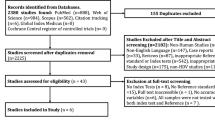
We identified 272 published studies, of which 23 studies met the inclusion criteria for prevalence assessment, representing a total of 11,467 people ( Fig 1 and Table 2 ). Three of these studies also screened individuals for occult HBV infection (OBI) in a total of 666 people using HBV DNA polymerase chain reaction (PCR) in addition to testing for HBsAg seroprevalence. Two studies screened initially with HBsAg, then with HBV DNA PCR on those who were HBsAg negative [ 20 , 21 ]. A further study included two different populations: a) those attending a clinic for sex workers, whom they screened initially for HBsAg, then HBV PCR in those who were HBsAg negative and b) known HBsAg negative, jaundiced participants whom they screened with HBV DNA PCR to detect OBI [ 22 ].
(AJOL: African Journal Online). All eight studies included for genetic analysis contain information on HBV genotype. Figure created in Biorender.com with licence to publish.
https://doi.org/10.1371/journal.pgph.0001165.g001
https://doi.org/10.1371/journal.pgph.0001165.t002
We identified nine studies reporting HBV sequence data (full or partial genome), including seven studies from among the 23 seroprevalence studies described above ( Table 2 ), and an additional two studies that only included HBsAg positive participants so were not included in prevalence analysis [ 23 , 24 ]. One study did not clearly report how many HBV samples were sequenced or the genotyping results, and this study was excluded from further analysis [ 25 ]. Eight studies remained for analysis representing 247 individuals ( Table 2 ).
We identified eight studies reporting HBsAg prevalence in low-risk populations (total number of individuals = 6828), seven studies in people living with HIV (medium risk, total number of individuals = 1861), five studies in high-risk groups (total number of individuals = 2221) and three studies in people presenting to clinical services with established liver disease (defined here as very high-risk for HBV infection; total number of individuals = 492).
(ii) Geographical distribution of HBV seroprevalence data
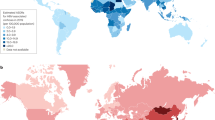
Of the 23 studies included, 14 (61%) were in Nairobi or Mombasa, Kenya’s most populous cities ( Table 2 ), and all studies were done in the South of the country along the infrastructure routes between Mombasa, Nairobi and Kisumu. These are also the most densely populated Kenyan counties [ 48 ]. Kisumu was the city most represented in the studies by overall sample size ( Fig 2 ).
Data from a systematic review of papers reporting prevalence and genetic data for HBV in Kenya between 2000 and 2021. The size of the red circle indicates numbers screened in each location, studies in the same location are grouped together. n = number of individuals reported. Surrounding countries are marked in blue, Kenya’s four most populous cities are marked in black. Figure created using R version 4.2.0, packages ggmaps version 3.0.0, ggplot2 version 3.3.6 and sf version 1.0–7. The Kenyan county shapefiles were obtained from the Humanitarian Data Exchange, available open source from https://data.humdata.org/dataset/geoboundaries-admin-boundaries-for-kenya .
https://doi.org/10.1371/journal.pgph.0001165.g002
The mean cohort sample size was 599 participants (IQR 434). 14 studies recruited participants for cohort inclusion at outpatient clinics (8 in HIV clinics, 4 in blood donor clinics, 1 in a health clinic and 1 in antenatal clinic), one captured data through the blood donor registry, three undertook community outreach screening, three recruited hospital inpatients, one recruited healthcare workers and one was a national survey of urban and rural population groups ( Table 2 ).
(iii) Quality assessment of the literature
Overall the quality of studies investigating HBV prevalence in Kenya was low ( Fig 3 and S1 Table ). 17/23 studies were cross sectional, reporting HBV population prevalence at a single time point only. Most cohort sampling methods were non-randomised and only 4/21 studies detailed their sample size calculation [ 20 , 21 , 37 , 41 ]. Several studies sampled people only from small geographical locations or from a subset of the general population e.g. HIV negative individuals. 21/23 studies used either an enzyme linked immunosorbent assay or chemiluminescent enzyme immunoassay (ELISA or CLEIA) for HBsAg diagnosis. Two studies used reverse passive haemagglutination for diagnosis of CHB, a method previously demonstrated to have poor sensitivity [ 26 , 27 , 43 ] ( Table 2 ). 2/23 studies went on to screen the HBsAg negative population for HBV DNA via PCR [ 20 , 21 ] and one study included a known HBsAg negative population which they screened for HBV DNA [ 22 ].
This is stratified by number of participants, study design, sampling method, data collection and diagnostic methods. RCT: Randomised controlled trial; EIA: Chemiluminescent enzyme immunoassay; ELISA: Enzyme linked immunosorbent assay.
https://doi.org/10.1371/journal.pgph.0001165.g003
(v) HBV prevalence estimates in different risk groups
The pooled estimate for HBV prevalence using a random effects model in the low-risk group was 3.36% (95% CI 2.67–4.21%) compared with 6.14% in the moderate risk group (95% CI 5.08–7.41%), 6.18% (95% CI 4.6–8.19%) in the high-risk group and 29.19% (95% CI 12.15–55.14%) in the very high-risk group, however we note that the confidence interval of this estimate is very wide ( Fig 4 ). Heterogeneity was significant (I 2 > 50%) within each subgroup, and highest in the very high-risk sub-group (I 2 = 95%, p < 0.01).
Data generated through a systematic review reporting prevalence and genetic data for HBV in Kenya between 2000–2021. In each case, the size of the population included is represented by the size of the square. Point prevalence and 95% Confidence Interval (CI) is indicated for each study. Studies are ordered by HBV prevalence in each risk group.
https://doi.org/10.1371/journal.pgph.0001165.g004
Three studies screened for OBI using HBV DNA PCR. These were in populations known to be HBsAg negative and from different HBV risk groups: blood donors, those living with HIV and those presenting to hospital with jaundice. OBI prevalence estimates in these studies were 2.4%, 5.3% and 18.7% respectively [ 20 – 22 ].
(vi) Identification of HBV sequences
All eight studies including HBV genetic information used PCR of the HBV basal core promotor, Pol or S genes for amplification, followed by Sanger sequencing to determine genotype. Two studies looked for known drug resistance-associated mutations (RAMs) [ 23 , 24 ]. Two studies undertook whole genome HBV sequencing in a total of 22 patients [ 23 , 28 ]. 228/247 (92%) of participants were infected with HBV genotype A, 15/247 (6%) with genotype D infection, whilst the remaining were either mixed genotype populations (2/247) or genotype D/E recombinants (2/247) ( Table 3 ). Sub-genotype was determined in 146/247 (59%) participants. This was most commonly sub-genotype A1 (134/146, 92%) in keeping with previous regional data [ 44 ].
Data from 8 studies marked * in Table 2 .
https://doi.org/10.1371/journal.pgph.0001165.t003
To provide further background context for HBV sequences in Kenya, we identified 25 full length HBV sequences from GenBank ( Fig 5 ). These were generated from three studies, published in 2013, 2015 and 2016 [ 24 , 28 , 45 ]. They primarily represented individuals presenting to hospital with jaundice (21/25 sequences), infected with genotypes A1 and D.
Kenyan sequences are those published in GenBank (downloaded 1 st Dec 2021) and are shown in red alongside genotype reference sequences in black (1000 bootstrap replicates were performed, and bootstrap support of ≥70% are indicated. Reference sequences from McNaughton et al. (2020) [ 11 ].
https://doi.org/10.1371/journal.pgph.0001165.g005
5/8 studies provided a detailed analysis of either amino acid or nucleotide substitutions found in the sequenced region of HBV [ 20 , 23 , 24 , 33 , 37 ]. 2/5 studies correlated these with known drug resistance mutations to lamivudine and other nucleoside analogues ( Table 4 ) [ 20 , 33 ]. One study reported the emergence of drug resistance mutations during lamivudine treatment associated with breakthrough HBV viraemia [ 33 ]. Multiple other mutations were described in the five studies, some of which were in the major hydrophilic region of the surface gene, and thus potentially important in influencing both natural and vaccine-mediated immunity [ 46 , 47 ].
https://doi.org/10.1371/journal.pgph.0001165.t004
(vi) HBV serology and HBV biomarkers
Exploring the prevalence of anti-HBs (vaccination or exposure) and anti-HBc (exposure to HBV) in HBsAg-negative populations is important to build up a full picture of population epidemiology. Among individuals testing HBsAg-positive, a panel of biomarkers is used to determine treatment eligibility, including HBeAg status, HBV DNA viral load, liver enzymes and imaging scores. These parameters are outside the primary scope of this study, but the data can be accessed as a supporting data file [ 48 ].
Enhanced efforts to characterise the epidemiology and disease burden of HBV are urgently required in Africa, as HBV is present at medium to high endemicity in many populations but has been neglected as a public health problem. Here we have reviewed the literature available on prevalence, genotypes and drug resistance data for CHB in Kenya. In our ‘low-risk’ category, intended to provide estimates most reflective of the general population, the pooled prevalence estimate for HBV infection was 3.4%. Point-prevalence estimates of ~6% were obtained for the groups we defined as medium and high risk, comprising people living with HIV infection and those with other identified risk factors for blood-borne virus infection. Similar prevalence estimates in the moderate- and high-risk groups was only evident after analysis. The number of studies was too low to allow for further subdivision into individual risk groups (e.g. comparing people who inject drugs, MSM, and sex workers). In the population presenting to healthcare facilities with established symptomatic liver disease (classified here as ‘very high risk’), the prevalence of HBV was 29.2% (although the underlying primary risk factor(s) for HBV acquisition in this group are not established).
In this very high-risk group, wide confidence intervals along with significant heterogeneity (I 2 = 95%) are notable. This population evidently has very different pre-test probabilities for HBV infection depending on underlying risk factors. In the absence of robust screening programmes, many people do not find out they have HBV infection until presenting to hospital with manifestations of liver disease. While the prevalence in this group evidently cannot be extrapolated to the general population, it is nevertheless an important observation that HBV in this setting accounts for such a high proportion of end-stage liver disease. Furthermore 2/3 studies in this very high-risk group used RPHA for HBsAg detection which is less sensitive than HBsAg, and therefore may underestimate true prevalence of HBV infection.
Most studies included in this review focussed on specific groups of people such as blood donors and those co-infected with HIV. Blood donation in Kenya is voluntary and often done by family members of those in need. There is no financial compensation for donation [ 49 ]. Routine screening for HBV through the Kenyan National Blood Transfusion Service (KNBTS) consists of ELISA for HBsAg only and there is no nucleic acid amplification testing (NAAT); some OBI may therefore go unidentified. Only one study in this review focussed on pregnant women [ 37 ] and one study enrolled healthcare workers [ 41 ]. These are accessible and important groups to screen for HBV infection given they are engaged with healthcare, likely to come for follow up visits, and interventions can have a significant impact on reducing transmission events. Treatment for pregnant mothers and healthcare workers would reduce onward transmission, and vaccination uninfected healthcare workers and babies at birth would decrease the overall burden of infection, reducing morbidity and mortality. One study was nationwide [ 38 ], but only included those who were HIV negative. More general population screening is lacking, and testing is not routinely done when presenting to healthcare facilities [ 50 ]. Some areas of Kenya have been more rigorous in their diagnostic approaches, but this is sporadic and may be increased only when there is a known outbreak of HBV in the local community, as has been the case in other African countries [ 51 , 52 ]. This may give a skewed view on population prevalence, but also leads to missed opportunities for diagnosis and intervention, particularly given the very high proportion of those presenting to hospital with jaundice or hepatitis found to be infected with HBV (pooled HBV prevalence 29.19% and 18.7% OBI prevalence).
It is notable that no studies were done in Northern Kenya, particularly along the borders with Somalia and South Sudan where the prevalence of HBV is likely to be substantially higher (for these two neighbouring countries, HBsAg prevalence is estimated at 19% and 12% respectively [ 53 , 54 ], however population density here is also very low [ 55 ].
Along with minimal population screening, there is very little sequencing of HBV in Kenya. Among the 25 papers we reviewed regarding HBV sequencing, only two reported whole genome sequencing, and none did next generation sequencing. We identified only 25 complete HBV genomes from Kenya in a GenBank search. Most available data is from single gene PCR and Sanger sequencing of S and P genes to determine genotype. Expanding these data will allow identification of recombinant genotypes, of which there is evidence in Kenya [ 28 , 31 ], but currently without good understanding of how these translate into clinical outcomes. Deep sequencing data will enable detection of minority variant mutations that may be relevant in emergence of vaccine escape and drug resistance, and also allow description of viral quasispecies, how this correlates with clinical phenotype and other biomarkers.
Three studies reviewed here screened for OBI using PCR. OBI prevalence was similar to estimated pooled HBsAg prevalence in the associated risk group (2.4%, 5.3% and 18.7% OBI prevalence in low, medium and high/very-risk groups compared with 3.36%, 6.14% and 6.18/29.19% pooled HBsAg positivity estimates in the equivalent groups). This indicates that many HBV cases are being missed due to the lack of appropriate screening tests, however the cost and poor availability of HBV DNA testing means it is not currently feasible to use as a universal screening test in Kenya. 20/23 studies solely reported HBsAg positivity diagnosed using other less sensitive tests. It is worth noting that of those presenting to hospital with jaundice who were HBsAg negative, nearly 20% were HBV DNA positive. It is not known whether the jaundice was due to acute HBV infection, or reactivation of chronic disease, but it seems to be an important indicator of HBV infection and screening of all those presenting to hospital with jaundice or hepatitis for OBI with HBV DNA PCR would be optimal. Few studies had characterised HBV exposure and vaccination status using anti-HBc and anti-HBs respectively. This highlights a broader issue around funding and access to laboratory tests needed for complete epidemiological assessment of populations.
HIV coinfection as a special case
The prevalence of HIV infection in adults in Kenya is 4.2% (95% CI 3.7–4.9%) [ 56 ]. Seven studies included in this analysis reported HBV prevalence in people living with HIV. The pooled HBV prevalence in this group was 6.14% (95% CI 5.08–7.41%). The HIV population is better represented than other groups at risk, as HBV screening is easier to offer to individuals already accessing healthcare for HIV monitoring and treatment. Through this established infrastructure for HIV (including clinics with staff, laboratory support, blood monitoring and drug distribution services), clinical care pathways for HBV could be incorporated. Although tenofovir is available free of charge in Kenya and is on the WHO list of essential medicines [ 57 ], it is only consistently available in combination with lamivudine or emtricitabine for HIV treatment, leaving the HBV monoinfected population unable to access licensed monotherapy.
Limitations
The HBV prevalence estimates we have generated here are wide and vary significantly between the risk groups (pooled risk group prevalence 3.36% - 29.19%). The very high-risk group also has a very wide confidence interval for prevalence estimates. Our risk groups were determined a priori based on existing understanding of the distribution of HBV infection, but data were insufficient to disaggregate into more specific groups, and we recognise that the prevalence of HBV infection in populations at risk varies substantially by setting. Other sources have different estimates of Kenyan HBV prevalence (e.g. 1% by the CDA Foundation [ 4 ]). The CDA data are from 2016, so may be out of date, but the varying estimates reflect difficulties with methods of data collection, varying data sources and data missingness. The overall quality of studies was low, with non-random sample selection common, no calculation of sample size in most studies and nearly all studies being cross sectional representing only a snapshot of HBV prevalence. Only selected populations are represented by the studies we identified, and even those studies seeming to represent the population more broadly are subject to bias. For example, the study of healthcare workers was primarily female nurses [ 41 ] and the nationwide survey only included HIV negative participants [ 38 ]. We considered only including those studies reaching a certain quality threshold in the prevalence meta-analysis, however this would have substantially restricted the available data. For example, including only those studies with random sampling methods and a documented sample size calculation would have left only three studies. One of the key findings of this systematic review is the lack of good quality seroprevalence data, and detailing this gives a good understanding of available literature.
There are no data for the northern part of Kenya, including the region around the border with South Sudan where there might be migration of high prevalence populations. It is likely that prevalence of HBV infection varies significantly by age, region of the country, and according to particular at-risk groups–thus targeted surveillance is important to provide an evidence-base for local and population-specific interventions.
No children were included in this review. In 2019 Kenya achieved an average coverage of 91% of 3 rd dose HBV childhood vaccination [ 58 ], but in future studies, screening children for HBsAg, anti-HBc and anti-HBs by birth cohort would be important to determine the impact of the vaccine campaign on infection, exposure and immunity, and to identify any populations being missed by vaccine coverage. There are increasing calls for the scale-up of BD HBV immunisation as part of a triple elimination campaign.
We highlight the poor representation of HBV in Kenya with sequencing data, identifying only two studies that undertook whole genome sequencing. 24/25 sequences available on GenBank were from two studies. This is clearly not representative of HBV in the general population, and work is required to determine circulating genotypes and to characterise polymorphisms that are relevant to outcomes of infection, treatment and vaccination.
Conclusions
We have assimilated epidemiological data for HBV in Kenya, together with genetic parameters where available, to provide the most refined picture possible to date. Our data suggest that Kenya falls into the ‘intermediate’ prevalence group (2–5%, as defined by the WHO). A sparse literature highlights the pressing need for clinical and research enterprise, to provide an evidence base for realistic and practical strategies that support country-specific scale-up of screening and treatment. Alongside continued efforts for three-dose vaccine coverage in infancy, enhanced interventions may include focus on HBV birth dose vaccine as part of the triple elimination initiative, with improved access to diagnostics, surveillance and treatment, to curtail the burden of disease in those currently infected, and reduce the incidence of new infections, moving Kenya towards 2030 elimination targets.
Supporting information
S1 checklist. preferred reporting items for systematic review and meta-analysis (prisma) 2020 statement checklist..
https://doi.org/10.1371/journal.pgph.0001165.s001
S1 Table. Joanna Briggs critical appraisal checklist.
https://doi.org/10.1371/journal.pgph.0001165.s002
Acknowledgments
This manuscript was written with the permission of the Director, KEMRI-CGMRC.
- 1. World Health Organisation. Global hepatitis report, 2017 [Internet]. 2017 [cited 2021 Nov 10]. https://www.who.int/publications/i/item/global-hepatitis-report-2017
- View Article
- PubMed/NCBI
- Google Scholar
- 3. WHO. Global health sector strategy on viral hepatitis 2016–2021. Global Hepatitis Programme Department of HIV/AIDS [Internet]. 2016 [cited 2021 Nov 9];(June):56. https://www.who.int/publications/i/item/WHO-HIV-2016.06
- 4. Countries Dashboard–CDA Foundation [Internet]. [cited 2022 Feb 7]. https://cdafound.org/polaris-countries-dashboard/
- 8. Kafeero HM, Ndagire D, Ocama P, Kudamba A, Walusansa A, Sendagire H. Prevalence and predictors of hepatitis B virus (HBV) infection in east Africa: evidence from a systematic review and meta-analysis of epidemiological studies published from 2005 to 2020. Archives of Public Health. 2021 Dec 1;79(1).
- 9. Ministry of Health. Kenya, National Policy Guidelines on Immunization 2013. 2013. 20 p.
- 10. Triple elimination initiative of mother-to-child transmission of HIV, syphilis and hepatitis B [Internet]. [cited 2022 Sep 12]. https://www.who.int/initiatives/triple-elimination-initiative-of-mother-to-child-transmission-of-hiv-syphilis-and-hepatitis-b
- 16. PRISMA 2020 Checklist Section and Topic Item # Checklist item Location where item is reported TITLE Title 1 Identify the report as a systematic review. [cited 2022 Feb 16]; http://www.prisma-statement.org/
- 17. critical-appraisal-tools—Critical Appraisal Tools | Joanna Briggs Institute [Internet]. [cited 2022 Feb 9]. https://jbi.global/critical-appraisal-tools
- 48. Downs L, Campbell C, Githinji G, Ansari A, Matthews P, Etyang AO. Treatment Eligibility Criteria Assessment in Hepatitis B Virus (HBV) Prevalence Studies in Kenya. 2022 Sep 12 [cited 2022 Sep 12]; /articles/journal_contribution/Treatment_Eligibility_Criteria_Assessment_in_Hepatitis_B_Virus_HBV_Prevalence_Studies_in_Kenya_/21063880/2
- 49. CDC Global Health—Kenya—Blog: Giving Blood, Giving Life [Internet]. [cited 2022 Feb 8]. https://www.cdc.gov/globalhealth/countries/kenya/blog/giving.htm
- 51. Nosocomial Outbreak of Hepatitis B Virus Infection in a Pediatric Hematology and Oncology Unit in South Africa: Epidemiological Investigation and Measures to Prevent Further Transmission. 2015;
- 55. 2019 Kenya Population and Housing Census Volume I: Population by County and Sub-County—Kenya National Bureau of Statistics [Internet]. [cited 2022 Mar 15]. https://www.knbs.or.ke/?wpdmpro=2019-kenya-population-and-housing-census-volume-i-population-by-county-and-sub-county
- 56. Kenya | UNAIDS [Internet]. [cited 2022 Feb 22]. https://www.unaids.org/en/regionscountries/countries/kenya
- 57. eEML—Electronic Essential Medicines List [Internet]. [cited 2022 Feb 22]. https://list.essentialmeds.org/?query=tenofovir%20disoproxil%20fumarate
- 58. Hepatitis B (HepB3) immunization coverage among 1-year-olds (%) [Internet]. [cited 2022 Feb 16]. https://www.who.int/data/gho/data/indicators/indicator-details/GHO/hepatitis-b-(hepb3)-immunization-coverage-among-1-year-olds-(-)
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
- My Bibliography
- Collections
- Citation manager
Save citation to file
Email citation, add to collections.
- Create a new collection
- Add to an existing collection
Add to My Bibliography
Your saved search, create a file for external citation management software, your rss feed.
- Search in PubMed
- Search in NLM Catalog
- Add to Search
Current topics in hepatitis B
Affiliation.
- 1 Academic Centre for Travel Medicine and Vaccines, WHO Collaborating Centre for Reference and Research on Viral Diseases, Royal Free Campus, Royal Free and University College Medical School, London NW3 2PF, UK.
- PMID: 11023756
- DOI: 10.1053/jinf.2000.0720
Over two billion people around the world have been infected with hepatitis B virus, of whom over 350 million are chronic carriers. Some 25% of carriers develop progressive liver disease. The annual mortality from hepatitis B infection and its sequelae is 1-2 million people worldwide.The following current topics are reviewed: immunization strategies against hepatitis B and the kinetics and antibody response; the controversy on screening blood donors for anti-core antibodies; mutations of hepatitis B surface antigen, including evidence that not all such mutants are detectable by current laboratory tests and, finally, the introduction of second generation nucleoside analogues for treatment of chronic hepatitis B infection, including treatment of patients with decompensated liver disease and liver transplantation.
Copyright 2000 The British Infection Society.
PubMed Disclaimer
Similar articles
- Hepatitis B virus infection. Shayeb J, Soweid A. Shayeb J, et al. J Med Liban. 2001 Mar-Apr;49(2):66-70. J Med Liban. 2001. PMID: 11910969 Review. No abstract available.
- A multi-center open study to determine the effect of lamivudine on HBV DNA clearance and to assess the safety of the regimen in patients with chronic hepatitis B infection. Mazur W, Król F, Cianciara J, Nazzal K, Gładysz A, Juszczyk J, Bolewska B, Adamek J, Czajka B, Swietek K, Kryczka W, Gonciarz Z. Mazur W, et al. Med Sci Monit. 2002 Apr;8(4):CR257-62. Med Sci Monit. 2002. PMID: 11951067 Clinical Trial.
- [Chronic hepatitis B. Recent advances in diagnosis and treatment]. Bernardi M, Biselli M, Gramenzi A. Bernardi M, et al. Recenti Prog Med. 2002 Jul-Aug;93(7-8):397-402. Recenti Prog Med. 2002. PMID: 12138683 Review. Italian.
- Clinical impact and efficacy of lamivudine therapy in de novo hepatitis B infection after liver transplantation. Castells L, Vargas V, Rodríguez F, Allende H, Buti M, Sánchez-Avila JF, Jardí R, Margarit C, Pumarola T, Esteban R, Guardia J. Castells L, et al. Liver Transpl. 2002 Oct;8(10):892-900. doi: 10.1053/jlts.2002.35555. Liver Transpl. 2002. PMID: 12360430
- High-titer antibody to hepatitis B surface antigen before liver transplantation can prevent de novo hepatitis B infection. Su WJ, Ho MC, Ni YH, Chen HL, Hu RH, Wu YM, Chang MH, Lee PH. Su WJ, et al. J Pediatr Gastroenterol Nutr. 2009 Feb;48(2):203-8. doi: 10.1097/MPG.0b013e3181819ad4. J Pediatr Gastroenterol Nutr. 2009. PMID: 19179883
- Prevalence of occult hepatitis B virus infection in Egypt: a systematic review with meta-analysis. Azzam A, Khaled H, El-Kayal ES, Gad FA, Omar S. Azzam A, et al. J Egypt Public Health Assoc. 2023 Jul 26;98(1):13. doi: 10.1186/s42506-023-00138-4. J Egypt Public Health Assoc. 2023. PMID: 37491501 Free PMC article. Review.
- Seroprevalence of hepatitis B virus surface antigen (HBsAg) in Egypt (2000-2022): a systematic review with meta-analysis. Azzam A, Khaled H, Elbohy OA, Mohamed SA, Mohamed SMH, Abdelkader AH, Ezzat AA, Elmowafy AOI, El-Emam OA, Awadalla M, Refaey N, Rizk SMA. Azzam A, et al. BMC Infect Dis. 2023 Mar 10;23(1):151. doi: 10.1186/s12879-023-08110-5. BMC Infect Dis. 2023. PMID: 36899311 Free PMC article.
- Demographics and Epidemiology of Hepatitis B in the State of Qatar: A Five-Year Surveillance-Based Incidence Study. Al Romaihi HE, Ganesan N, Farag EA, Smatti MK, Nasrallah GK, Himatt SM, Derbala MF, Alshamali M, Mahadoon LK, Khogali HS, Sallam M, Al Thani AA, Al Thani M, Al Kaabi S, Yassine HM. Al Romaihi HE, et al. Pathogens. 2019 May 21;8(2):68. doi: 10.3390/pathogens8020068. Pathogens. 2019. PMID: 31117254 Free PMC article.
- Association of IL-10 and IL-10RA single nucleotide polymorphisms with the responsiveness to HBV vaccination in Chinese infants of HBsAg(+)/HBeAg(-) mothers: a nested case-control study. Wen S, Wu Y, Pan Y, Cao M, Zhao D, Wang C, Wang C, Kong F, Li J, Niu J, Jiang J. Wen S, et al. BMJ Open. 2018 Nov 28;8(11):e022334. doi: 10.1136/bmjopen-2018-022334. BMJ Open. 2018. PMID: 30498038 Free PMC article.
- Hepatitis B virus genotypes among chronic hepatitis B patients reporting at Korle-Bu teaching hospital, Accra, Ghana. Dongdem AZ, Dzodzomenyo M, Asmah RH, Nyarko KM, Nortey P, Agyei A, Adjei DN, Kenu E, Adjei AA. Dongdem AZ, et al. Pan Afr Med J. 2016 Oct 1;25(Suppl 1):5. doi: 10.11604/pamj.supp.2016.25.1.6170. eCollection 2016. Pan Afr Med J. 2016. PMID: 28210373 Free PMC article.
Publication types
- Search in MeSH
Related information
- PubChem Compound (MeSH Keyword)
LinkOut - more resources
Full text sources.
- Elsevier Science
- MedlinePlus Health Information
- Citation Manager
NCBI Literature Resources
MeSH PMC Bookshelf Disclaimer
The PubMed wordmark and PubMed logo are registered trademarks of the U.S. Department of Health and Human Services (HHS). Unauthorized use of these marks is strictly prohibited.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Published: 20 July 2022

A roadmap for serum biomarkers for hepatitis B virus: current status and future outlook
- Anna Kramvis 1 ,
- Kyong-Mi Chang ORCID: orcid.org/0000-0001-6811-9364 2 ,
- Maura Dandri 3 , 4 ,
- Patrizia Farci 5 ,
- Dieter Glebe ORCID: orcid.org/0000-0001-5039-0252 6 , 7 ,
- Jianming Hu 8 ,
- Harry L. A. Janssen 9 ,
- Daryl T. Y. Lau 10 ,
- Capucine Penicaud ORCID: orcid.org/0000-0002-2708-8773 11 ,
- Teresa Pollicino ORCID: orcid.org/0000-0001-6602-3035 12 ,
- Barbara Testoni ORCID: orcid.org/0000-0001-5588-5465 13 , 14 ,
- Florian Van Bömmel 15 ,
- Ourania Andrisani 16 ,
- Maria Beumont-Mauviel 17 ,
- Timothy M. Block 18 ,
- Henry L. Y. Chan 19 , 20 ,
- Gavin A. Cloherty 21 ,
- William E. Delaney 22 ,
- Anna Maria Geretti 23 , 24 , 25 ,
- Adam Gehring ORCID: orcid.org/0000-0003-1150-5840 26 ,
- Kathy Jackson 27 ,
- Oliver Lenz 28 ,
- Mala K. Maini ORCID: orcid.org/0000-0001-6384-1462 29 ,
- Veronica Miller 30 ,
- Ulrike Protzer ORCID: orcid.org/0000-0002-9421-1911 31 ,
- Jenny C. Yang 32 ,
- Man-Fung Yuen 33 , 34 ,
- Fabien Zoulim ORCID: orcid.org/0000-0002-2245-0083 35 &
- Peter A. Revill ORCID: orcid.org/0000-0003-2715-2541 27 , 36
Nature Reviews Gastroenterology & Hepatology volume 19 , pages 727–745 ( 2022 ) Cite this article
25k Accesses
64 Citations
123 Altmetric
Metrics details
- Diagnostic markers
- Prognostic markers
Globally, 296 million people are infected with hepatitis B virus (HBV), and approximately one million people die annually from HBV-related causes, including liver cancer. Although there is a preventative vaccine and antiviral therapies suppressing HBV replication, there is no cure. Intensive efforts are under way to develop curative HBV therapies. Currently, only a few biomarkers are available for monitoring or predicting HBV disease progression and treatment response. As new therapies become available, new biomarkers to monitor viral and host responses are urgently needed. In October 2020, the International Coalition to Eliminate Hepatitis B Virus (ICE-HBV) held a virtual and interactive workshop on HBV biomarkers endorsed by the International HBV Meeting. Various stakeholders from academia, clinical practice and the pharmaceutical industry, with complementary expertise, presented and participated in panel discussions. The clinical utility of both classic and emerging viral and immunological serum biomarkers with respect to the course of infection, disease progression, and response to current and emerging treatments was appraised. The latest advances were discussed, and knowledge gaps in understanding and interpretation of HBV biomarkers were identified. This Roadmap summarizes the strengths, weaknesses, opportunities and challenges of HBV biomarkers.
As new therapies for hepatitis B virus (HBV) infection become available, new biomarkers to monitor viral and host responses are urgently needed.
This Roadmap summarizes current knowledge on existing and emerging serum biomarkers in the context of chronic HBV infection.
This Roadmap discusses the strengths, weaknesses, opportunities and challenges of serum HBV biomarkers.
This Roadmap provides suggestions of the way forward to advance the biomarkers required to fast-track an HBV cure for all, irrespective of resources, HBV genotype or disease stage.
Similar content being viewed by others
Global burden of hepatitis B virus: current status, missed opportunities and a call for action
Assessing the diagnostic accuracy of serological tests for hepatitis delta virus diagnosis: a systematic review and meta-analysis
Breakthroughs in hepatitis C research: from discovery to cure
Introduction.
Hepatitis B virus (HBV) infection can cause chronic hepatitis B (CHB), which can result in severe liver disease, including cirrhosis and liver cancer. A major challenge to recovery, even in treated individuals, is the persistence of two forms of the viral genome in hepatocytes: the replication-competent, episomal, covalently closed circular DNA (cccDNA), and the linear subgenomic HBV sequences integrated into the human genome, which do not sustain viral replication but can express some HBV antigens 1 . High viral loads and antigens can lead to T and/or B cell exhaustion and downregulation of innate immune sensors and pathways 2 , 3 , 4 , 5 , 6 , 7 . Current antiviral therapies, which include nucleos(t)ide analogues (NUCs) and pegylated interferon-α (peg-IFNα), decrease viral loads and lead to remission of the disease. However, although NUCs are well tolerated, they require lifelong treatment and do not target cccDNA directly 8 . Conversely, peg-IFNα, the only finite treatment for CHB, is less well tolerated but might affect cccDNA directly and indirectly 9 . Treatment results in hepatitis B surface antigen (HBsAg) loss (also known as functional cure) in a minority of cases 10 , 11 . Consequently, new effective, finite and well-tolerated cure therapies are being sought to induce functional cure, fully controlling HBV replication and gene expression and/or ultimately eliminating cccDNA and integrated HBV DNA (also known as sterilizing cure) 10 , 11 .
CHB is a major global health challenge, and there is an urgent need to develop curative therapies for patients with CHB worldwide 12 . In 2020, mortality from human immunodeficiency virus (HIV) infection, malaria and tuberculosis continued to decline, but death attributable to viral hepatitis is still increasing 13 , with rates predicted to double by 2040, even though effective cures for hepatitis C virus are already available. The World Health Organization (WHO) set a goal for the elimination of viral hepatitis with a 90% reduction of new HBV cases by 2030; it is unlikely to be achieved without a substantial increase in the rate of HBV diagnosis. It is estimated that less than 10% of individuals with HBV infection have been identified, and only 10% of the eligible patients receive treatment globally 12 . To achieve the goal set by WHO, a panel of serum biomarkers will likely be required for surveillance to predict treatment response and outcome as an armamentarium of new therapies is developed. Although a limited number of biomarkers is available that permits monitoring of HBV DNA replication and treatment response to current treatment regimens, biomarkers accurately predicting functional cure are lacking. With more than 40 new therapeutic approaches in preclinical or clinical trials 14 , 15 targeting HBV replication or stimulating HBV-specific host immune responses, identifying suitable biomarkers will become increasingly important.
In October 2020, the International Coalition to Eliminate Hepatitis B Virus ( ICE-HBV ) held a virtual and interactive workshop on HBV biomarkers , at which stakeholders from academia, clinical practice and the pharmaceutical industry, with complementary expertise, presented and participated in panel discussions. The clinical utility of both classic and emerging, viral and immunological serum biomarkers with respect to the course of infection, disease progression, and response to current and emerging treatments was appraised. The latest advances were discussed and knowledge gaps in our understanding and interpretation of HBV biomarkers were identified.
This Roadmap summarizes current knowledge for existing and emerging HBV virological and immune-related biomarkers and suggests a road forward to advance the biomarkers required to fast-track an HBV cure for all, irrespective of resources, HBV genotype or disease stage.
HBV biomarkers
HBV cccDNA, the key molecule in the HBV life cycle, is first generated from incoming virions and exists as a stable minichromosome in non-dividing hepatocytes 1 , 16 . cccDNA is the template for transcription of all HBV RNAs 17 , including the pre-genomic RNA (pgRNA) replication intermediate that is reverse transcribed into new HBV genomes. Thus, cccDNA is responsible for the production of virions and subviral particles. A detailed description of the viral life cycle has been previously presented 18 . Integrated HBV sequences can encode HBsAg and seem to be a major source of HBsAg in patients who are negative for hepatitis B e antigen (HBeAg) 19 . In addition, integrated sequences can produce truncated HBV RNAs and hepatitis B virus x (HBx) protein.
Serum biomarkers currently used in clinical practice to discriminate CHB and disease stages 20 include quantitative HBsAg, HBeAg, HBV DNA (Fig. 1 ) and alanine aminotransferase (ALT) serum levels. However, these biomarkers are not universally available, particularly in resource-limited settings (discussed later), and the classification and use of these classic markers do not completely reflect CHB complexity or HBV intrahepatic activity 16 . Intrahepatic measurement of cccDNA and viral RNAs might improve disease classification but entail using liver biopsy samples, which are invasive, not routine CHB care and unavailable in resource-limited settings. Furthermore, only a small section of the liver is sampled by liver biopsies, and HBV is unevenly distributed in the liver 21 , 22 . Although specific quantitative polymerase chain reactions (PCRs) for cccDNA have been developed 23 , 24 , the coexistence of HBV replicative DNA intermediates in infected cells 16 , including relaxed circular and integrated HBV DNA molecules, interferes with accurate cccDNA quantification. In this respect, a global collaborative project initiated by ICE-HBV aims to optimize and harmonize cccDNA detection and quantification protocols in liver tissue and cell culture.
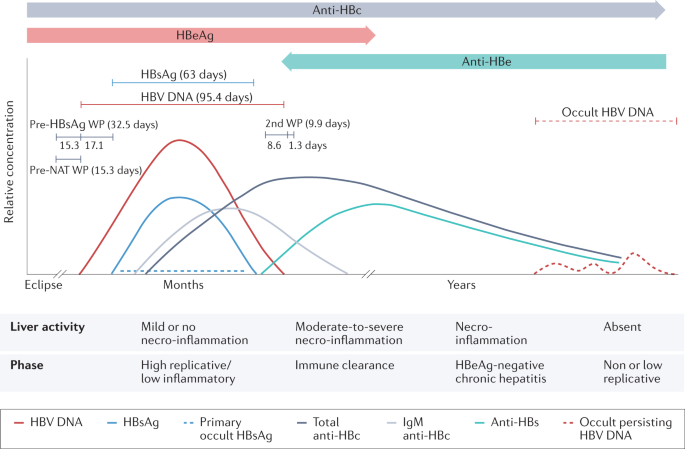
The curves in the upper part of the diagram show the relative concentration of the markers in a typical infection. The lines above the curves show the mean lengths of the detection periods of hepatitis B virus (HBV) DNA and hepatitis B surface antigen (HBsAg) as estimated from the numbers of HBV nucleic acid testing (NAT) yields, with and without detectable HBsAg. The lengths of the pre-HBsAg and post-HBsAg window periods (WPs) and pre-NAT and post-NAT WPs as described by Weusten et al. 176 . In a later stage of occult HBV infection, when titres of antibodies against hepatitis B surface antigen (anti-HBs) have declined to below 10–100 mIU/mL, occult persisting HBV DNA in the liver can reappear in plasma. If infection occurs perinatally or in very early childhood, there is no full recovery because of immune system immaturity, and this can lead to chronic infection in 90% of cases. The duration of HBsAg positivity is thus prolonged. The lower panel of the figure depicts the stages of natural infection according to current European Association for the Study of the Liver (EASL) guidelines (hepatitis B e antigen (HBeAg)-positive or HBeAg-negative disease and/or infection) 177 . Anti-HBc, hepatitis B c antibody; Anti-HBe, hepatitis B e antibody; HBeAg, hepatitis B e antigen. Adapted with permission from ref. 178 , Wiley.
Accordingly, there is a pressing need for alternative biomarkers that not only accurately reflect the intrahepatic cccDNA pool and transcriptional activity 25 , 26 but also better characterize the different CHB disease stages and risk of complications, detect HBV integration, improve the determination of hepatocellular carcinoma (HCC) risk, and monitor immune status and response to therapy. For example, one study showed that HBV functional cure in 10 of 14 patients with genotype A HBV infection was associated with anti-HBsAg immune complex peaks that overlapped with ALT flares in serum levels 27 . This suggests the utility of hepatitis B surface antibody (anti-HBs, also known as HBsAb) immune complexes as a biomarker of functional cure and warrants further investigation in larger studies encompassing additional HBV genotypes.
Ideal biomarkers should be predictive (visible early and indicative of clinical outcome), highly specific and sensitive, HBV (sub)genotype agnostic, correlative with disease activity and severity, reflective of durable viral control, reproducible, non-invasive and accessible, rapid, simple, and inexpensive 28 . Biomarkers should also be accessible in resource-limited settings.
Various serum HBV markers have been proposed as surrogates for intrahepatic viral activity. These markers include the complete virion (HBV DNA, hepatitis B core antigen (HBcAg), HBsAg), subviral particles (with HBsAg), empty virus particles (with HBsAg and HBcAg but without HBV DNA or RNA), viral particles containing HBV RNA, and HBV core-related antigen (HBcrAg) consisting of the non-particulate HBeAg and the related precore protein that, like HBeAg, is also derived from the precore/core open reading frame 29 . We have appraised the clinical utility of both classic (HBV DNA, HBeAg and/or hepatitis B e antibody (anti-HBe), HBsAg, anti-HBs and hepatitis B core antibody (anti-HBc)) and emerging (HBV RNA, HBcrAg and HBsAg isoforms) biomarkers of HBV infection with respect to the course of infection, disease progression, and response to current and emerging treatments.
Classic biomarkers: needs and limitations
A summary of classic biomarkers is presented in Supplementary Table 1 . More sensitive DNA assays might be beneficial in identifying residual and fluctuating HBV levels 30 , 31 , predicting the risk of reactivation or severe outcomes following NUC treatment withdrawal 32 , assessing the effect of direct antiviral agents on DNA suppression, and accurately detecting occult HBV infection (OBI) 33 . In light of WHO recommendations that, in settings where antenatal HBV DNA testing is not available, HBeAg testing can be used to determine eligibility for tenofovir therapy to reduce the likelihood of mother-to-child HBV transmission 34 , there is a need for point-of-care (POC) HBeAg assays, particularly in resource-limited settings. A limitation of all HBsAg assays is that they do not differentiate between HBsAg derived from cccDNA and integrated HBV DNA because the protein derived from either source is identical 35 . More research is required to determine the usefulness of quantitative anti-HBs, anti-HBe and anti-HBc assays to better characterize the risk of HCC and reactivation of HBV infection following treatment discontinuation or immunosuppression. These markers are proving useful in detecting OBI (discussed later).
Point-of-care testing: an unmet need
WHO has developed a simplified HBV treatment cascade based on the biomarkers of HBsAg, ALT, presence of cirrhosis and HBV DNA levels 36 . Treatment eligibility requires appropriate screening and assessment for active disease. These tools include rapid diagnostic tests and ELISA for HBsAg, HBV DNA nucleic acid testing (NAT), ALT (liver panel), and fibrosis measurements such as transient elastography or aspartate aminotransferase-to-platelet ratio index (APRI) score. In addition, regular HCC surveillance with abdominal ultrasonography alongside, or not, serum analysis of α-fetoprotein (AFP) serum levels is essential.
Unfortunately, many of these tests are not readily available as required POC tests, particularly in resource-limited settings 37 . Their availability will be necessary to meet the ASSURED criteria 38 (Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free, and Deliverable to those that need them).
Rapid HBsAg screening tests, such as Determine HBsAg 2 (Alere Medical, Chiba-ken, Japan) and VIKIA HBsAg (bioMérieux SA, Marcy-l’Étoile, France), are available for POC screening. POC HBV DNA NAT platforms have been validated or are in development. In addition, fibrosis measurement by transient elastography (FibroScan) can be adopted for POC with a portable FibroScan; however, the availability of transient elastography is sparse in resource-limited settings 39 .
Rapid diagnostic tests for HBsAg for multiple HBV genotypes and subtypes 40 , with results available in 30 min, have been developed 41 . Notably, the sensitivity of HBsAg enzyme-linked immunosorbent assay (ELISA) and rapid diagnostic tests varies for different HBV genotypes 41 . Genetic variability in the S gene region of HBV can also affect diagnostic efficacy and specificity 42 , 43 . HBsAg ELISA tests that include multiple monoclonal antibodies in the capture phase, together with a polyclonal conjugate phase, are more accurate. HBsAg rapid diagnostic tests are generally less sensitive than lab-based ELISA tests. In our opinion, rapid diagnostic tests, such as Determine and VIKIA, are adequate for HBV screening but are not ideal for monitoring treatment response.
Dried blood spot (DBS) tests have numerous advantages compared to obtaining a standard blood sample; namely, the capillary finger-stick does not require trained health workers, high blood volumes, basic lab facilities, electric power, or a cold chain for transport and storage 44 . In 2017, WHO conditionally recommended the use of DBS specimens as an option for HBV DNA NAT in settings where there were no facilities or expertise to take venous blood specimens and for persons with poor venous access 36 . Meta-analysis of 12 studies from Europe (France, Denmark, Germany and Spain), Africa (Ethiopia, Congo, Egypt and Zambia), India and Mexico comparing the sensitivity and specificity of DBS versus serum samples for HBV DNA showed that DBS sensitivity ranged from 93% to 100% and specificity from 70% to 100% 44 . The limit of detection of the HBV DNA assays for serum samples ranged from 10 IU/ml to 100 IU/ml. HBV DNA detection limits from DBS specimens ranged from approximately 900 IU/ml to 4,000 IU/ml (ref. 44 ). Potential issues identified were the various lengths of storage before testing, ambient temperature variations, and the absence of manufacturer validation for the use of their assays with DBS samples or standardization of technical guidance. Many manufacturers and investigators have validated NAT using DBS by standardized procedures.
DBS have also been used in the HBcrAg assay, showing that HBcrAg correlated strongly with HBV DNA levels for genotypes A–E in individuals with high viral load, suggesting that DBS might be useful in resource-limited settings with limited access to NAT 45 . However, the assay used to measure HBcrAg, LUMIPULSE chemiluminescent enzyme immunoassay (Fujirebio, Tokyo, Japan), is currently not widely available.
Several POC or near POC NAT platforms have been developed for other blood-borne viruses, including the Xpert HCV Viral Load FS assay (Cepheid, USA), Genefrive (Manchester, UK) and the Alere (Abbott, USA). The GeneExpert platform (Cepheid, USA) is widely available across resource-limited settings where it is used routinely for tuberculosis diagnostics and HIV viral load monitoring. It is also being used to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Importantly, a GenExpert viral load assay has also been developed for HBV, which should be readily available in many resource-limited settings where GeneExpert machines have been placed to monitor other pathogens 46 , 47 . Fibrosis measurement by transient elastography can be adopted for POC with a portable FibroScan. FibroScan is the preferred method for cirrhosis and fibrosis assessment, with improved performance compared to APRI/FIB4 (ref. 48 ). Improving the availability of the portable FibroScan in resource-limited settings should be prioritized. It is important to note that hepatic inflammation and non-fasting states can falsely increase fibrosis scores with FibroScan, so the results need to be interpreted with care.
To establish a POC model for HBV management in resource-limited settings, standardized diagnostic assays with rapid diagnostic tests or DBS using currently available HBV DNA and HBsAg biomarkers are urgently required. Together with practical and effective guidelines for disease monitoring and therapy, this will assist in reaching the WHO goals of HBV elimination, particularly in resource-limited settings, where the HBV burden is highest.
Novel or emerging biomarkers
HBV cccDNA is the template for five viral transcripts: precore/core RNA, pgRNA, preS1 RNA, preS2/S RNA and X RNA 17 , 49 , 50 . pgRNA and precore RNA are over-length molecules of approximately 3.5-kb in size, and hence can only be transcribed from cccDNA 51 , 52 , 53 , 54 , 55 . Additional molecules transcribed from cccDNA include 5′ truncated RNAs with 3′ poly(A) tails 56 , 57 and HBx RNA 55 , 58 . Subgenomic RNAs encoding HBx or HBsAg can also be transcribed from integrated HBV DNA, which could be 5′ truncated with poly(A) tails. At least 15 splice variants of pgRNA 54 , 55 , 56 and two splice variants of preS2/S mRNA 17 , which arise from co-transcriptional processing, have been isolated from the supernatant of transfected cell lines or primary human hepatocytes 54 and patient sera 54 , 59 , 60 , 61 . The majority of splice variants identified to date encode the 5′ region of pgRNA and their contribution to pgRNA levels detected in RNA PCR assays requires investigation.
Viral RNA does not circulate freely 52 but is found in virus-like particles in serum (or supernatant of cultured cells) 51 , 56 , 62 . HBV RNA can also be found in capsid-antibody complexes 62 and naked capsids 51 , 62 . The secreted HBV RNA-containing viral particles have a similar buoyant density to HBV DNA-containing particles. However, they are produced at lower levels and, when reverse transcription is blocked, levels increase relative to HBV DNA-containing particles 53 . The quasispecies of serum and intrahepatic HBV RNA are similar and homologous to cccDNA 56 .
Various strategies for measuring HBV RNA are shown in Table 1 . Few comparisons of the different assays have been performed so far and no widely accepted (international) RNA standard is currently available 63 .
Clinical relevance of HBV RNA
The ratios of the different forms of HBV RNA and their importance during the different clinical phases or treatment responses are unknown. However, we consider that HBV RNA shows promise as a biomarker of treatment responses that are not predicted by serum HBV DNA levels using current NUC therapy in some settings. HBV RNA kinetics are predictors of response to treatment in patients who are HBeAg positive 64 . Even though HBV DNA levels decrease following NUC treatment, HBV pgRNA levels can remain relatively high, decreasing at a later stage 63 . This decoupling of pgRNA to HBV DNA levels can enable pgRNA to be used as a surrogate marker for cccDNA activity or cccDNA copies in the cell under NUC therapy 63 .
In the absence of therapeutic intervention, the level of HBV RNA in serum generally correlates closely with HBV DNA, albeit 1.5–2 logs lower 65 , 66 . The prognostic usefulness of HBV RNA in following the natural history of infection is uncertain and it has become clear, through the preparation of this Roadmap, that there is a paucity of data on its ability to predict liver-related complications, including cirrhosis or HCC. Serum HBV RNA and intrahepatic HBV RNA (primarily full-length encapsidated pgRNA 51 , 67 ) levels were lower in patients with inactive HBV than in patients with CHB who were either HBeAg positive or HBeAg negative 65 , 68 . In patients with CHB, HBV RNA levels varied according to HBeAg status (being higher in patients who were HBeAg positive), with liver inflammation, HBV genotype, and basal core promoter and/or precore mutants 24 , 53 , 66 , 69 . In the HBeAg-positive phase, serum HBV RNA levels showed a better correlation to serum HBV DNA levels than to either HBsAg or HBeAg 66 . This correlation seemed to be genotype dependent, with HBV RNA showing a strong correlation with HBV DNA levels for genotype A and with HBsAg levels for genotypes B and C; these associations were weakest for genotype D 66 . The correlation of HBV RNA with HBV DNA held during the HBeAg-negative phase, with genotype-specific correlations of HBV RNA levels only determined for genotypes A and D 66 . The weak correlation between HBV RNA levels and HBsAg in the HBeAg-negative phase is most likely due to the large proportion of HBsAg being expressed from integrated HBV DNA.
The decline of both full-length and subgenomic HBV RNA at 3 and 6 months after the initiation of NUC treatment was the strongest predictor of HBeAg seroconversion compared to other markers, including serum levels of HBV DNA, HBsAg, HBeAg, HBcrAg and ALT as well as sex, age and HBV genotype 64 , 70 . Together with HBcrAg, serum HBV RNA levels are also a prognostic biomarker for predicting ALT flares and the likelihood of HBV reactivation following cessation of NUC therapy in the absence of detectable HBV DNA 32 , 71 . In patients who were HBeAg positive, HBV RNA correlated strongly with HBcrAg levels but this was not observed in patients who were HBeAg negative 32 , 71 . Increased HBV RNA levels can also be a marker for viral relapse after NUC discontinuation 72 , 73 .
As serum pgRNA is derived exclusively from HBV cccDNA, its measurement can reflect cccDNA activity 1 . It might also serve as a surrogate marker to assess the target engagement of drugs affecting serum RNA levels by affecting RNA transcription, pgRNA stability and pgRNA packaging (that is, pegylated interferons, small interfering RNAs, antisense oligonucleotides, core protein assembly modulators (also known as capsid assembly modulators (CAMs)). Indeed, the CAM NVR 3–778 plus peg-IFNα but not the NUC entecavir lowered the concentration levels of HBV RNA in serum without causing substantial changes in cccDNA loads 71 , 74 , 75 . Peg-IFNα treatment reduced HBV RNA levels in the liver and serum of humanized mice, with good correlations between serum and intrahepatic pgRNA levels but not with cccDNA levels, as such pgRNA reduction mostly reflected the suppression of cccDNA activity 76 . In patients who are HBeAg positive, low HBV RNA levels can also help predict response, HBeAg loss and sustained virological control off-treatment after peg-IFNα and combined peg-IFNα–NUC therapy 52 , 77 . Although the relevance and correlation between viral RNA serum levels and liver damage still need clarification, serum HBV RNA could help define treatment end points 51 , 78 .
HBV RNA can be an addition to HBV DNA as a biomarker in some settings, particularly in predicting which patients will benefit most from treatment cessation. However, because the contributions of serum HBV RNA derived from cccDNA, integrated HBV DNA, or splice variants were unresolved and different quantitative methods were used 79 , the clinical and biological importance of serum HBV RNA levels should be interpreted with caution. There are also currently no HBV RNA standards available to validate and compare assays in different laboratories. The current Abbott HBV RNA assay has utilized WHO-approved DNA standards 32 , 63 and, until appropriate HBV RNA standards are developed and calibrated, WHO DNA standards will continue to be used where applicable. HBV RNA was undetectable by currently available assays in more than 50% of patients who were HBeAg negative and on long-term NUC therapy and could even be undetectable in patients with low HBV DNA levels (as in HBeAg-negative infection) 32 who had not received treatment, suggesting the sensitivity of detection needs improvement, especially with regards to possible cross-reaction with HBV DNA. However, HBV RNA could be detected in patients who were HBV DNA negative and was shown to be an accurate predictor for patients who might relapse following NUC treatment cessation 32 , demonstrating the promise of this biomarker in clinical settings.
Hepatitis B core-related antigen
HBcrAg, a composite antigen found in the blood of patients with HBV infection, has emerged as a potential marker to monitor intrahepatic cccDNA and its transcriptional activity, thereby defining new meaningful treatment end points 80 , 81 , 82 . HBcrAg components and their biogenesis are illustrated in Fig. 2 . Each HBcrAg component can have distinct functions and applications in reflecting intrahepatic viral activities 29 , 83 , varying between genotypes and individual patients 29 .
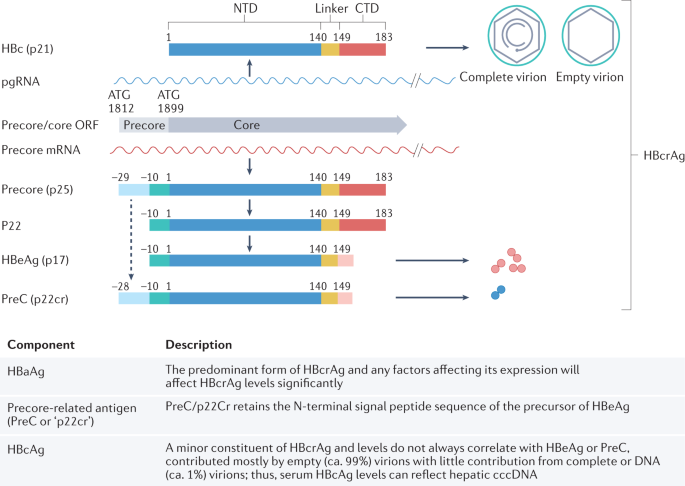
Hepatitis B core antigen (HBcAg), translated from pre-genomic RNA (pgRNA), forms the icosahedral capsid inside complete and empty virions 179 . The direct translation product from the precore mRNA is the precore precursor protein (p25), from which hepatitis B virus e antigen (HBeAg) and precore (PreC; also known as p22cr) are both derived. Removal of the N-terminal signal peptide of p25, by the signal peptidase during p25 translocation into the endoplasmic reticulum lumen, leads to the production of p22 (ref. 180 ), which is further processed at its C-terminal domain (CTD) before being secreted as the dimeric HBeAg (p17) 181 , 182 . cccDNA, covalently closed circular DNA; HBc, hepatitis B c; HBcrAg, hepatitis B virus core-related antigen; NTD, N-terminal domain; ORF, open reading frame. Adapted with permission from ref. 29 , American Society for Microbiology.
Clinical relevance of HBcrAg
HBcrAg can distinguish the different clinical phases of CHB 83 , 84 , although this ability is limited by the presence of basal core promoter and/or precore mutants that influence HBeAg levels. Serum HBcrAg levels were higher in the HBeAg-positive phase than in the HBeAg-negative phase 82 , 85 , 86 , and correlation with intrahepatic cccDNA levels and transcriptional activity was stronger during HBeAg-positive CHB 82 . HBcrAg also showed potential for distinguishing between HBeAg-negative inactive and active disease 82 , 85 , 87 , 88 , 89 , 90 , 91 . Using principal component analysis, researchers could discriminate between patients who were HBeAg positive or HBeAg negative. When HBcrAg was considered, a third group of patients was identified characterized by higher cccDNA, transcriptional activity, high fibrosis and necro-inflammatory activity that could not be discriminated by serum HBV DNA and HBeAg alone 32 , 82 , 92 . Multiple studies show that HBcrAg correlated well with pgRNA in HBeAg-negative CHB. An improved assay for HBcrAg assay with 10-fold increased sensitivity compared to previous assays has been developed 93 .
HBV DNA and pgRNA levels in the liver were higher in patients who were HBcrAg positive than in those who were HBcrAg negative, suggesting active HBV replication in HBcrAg-positive livers 94 . HBcrAg was a non-inferior biomarker to HBV DNA in predicting cirrhosis in patients who were HBeAg negative 95 , with elevated HBcrAg levels in patients with CHB who were treatment naive and HBeAg negative, correlating with increased risk of progression to cirrhosis 96 . Thus, although an elevated HBV DNA level is still the main indicator for initiation of NUC treatment, HBcrAg might also have a role in identifying patients of high risk with an intermediate HBV viral load who could benefit from early NUC treatment to prevent progression to cirrhosis 96 . HBcrAg levels can also predict the risk of HCC 82 , 94 , which is important as NUC therapy does not eliminate this risk 97 . In a large study of 2,666 patients with CHB who were infected with genotypes B or C, HBcrAg was an independent risk factor for HCC (209 patients were positive for HCC) 98 . Whether this observation applies to patients with different HBV genotypes requires investigation. Despite sustained viral suppression, persistently high levels of on-treatment HBcrAg and detectable levels after antiviral therapy termination might predict long-term HCC risk in patients with CHB treated with NUC 94 , 97 , 99 . Detection of residual HBcrAg, in combination with secreted HBV RNA but not HBV DNA, also predicted severe relapse following NUC treatment withdrawal in three of four patients 32 and might be a useful biomarker to contraindicate NUC re-treatment and predict relapse 100 following treatment cessation.
HBcrAg is also associated with response to current antiviral therapy for HBeAg-positive CHB 101 and, together with secreted HBV RNA, should be considered when evaluating new antiviral therapies aiming at a functional cure by direct or indirect targeting of the intrahepatic cccDNA activity 102 . Although used in Japan, further studies are needed to inform whether HBcrAg can be used in clinical practice more broadly.
HBcrAg assay
The current commercial HBcrAg assay, the chemiluminescent enzyme immunoassay (CLEIA; Lumipulse G HBcrAg, Fujirebio), is for research use only. It detects a combination of HBcAg, HBeAg (both free and in the HBeAg–HBe antibody complex) and precore proteins in the blood following sodium dodecyl sulfate and heat treatment 103 , 104 , 105 and has been validated for DBS 45 . The relative contribution of each component of HBcrAg in this assay has not been elucidated and affects the accuracy and utility of HBcrAg as a biomarker for cccDNA. Both viral and host factors can affect the expression of the different components of HBcrAg. Mutations that affect HBeAg (and precore) expression can influence HBcrAg levels, and this reduction obviously cannot be correlated to a reduction of cccDNA activity or copy number. This is also true for increased clearance of HBeAg (and precore) from the serum via a peripheral mechanism such as antibody-mediated clearance 29 , 106 .
HBsAg isoforms
The HBsAg components, large and medium surface proteins, differ during the various phases of CHB 107 and have different dynamics under treatment 107 , 108 . Except for genotype G, which has impaired HBsAg release, no difference in glycosylation, subcellular distribution, release of HBsAg or formation of subviral particles was evident between genotypes when compared in vitro 109 . However, there were differences in the proportions of HBsAg isoforms both intracellularly and extracellularly between different genotypes, with different post-translational modification patterns for large surface proteins 109 . Large and medium surface proteins were shown in patients to decrease earlier than small surface proteins prior to HBsAg loss, suggesting that these proteins might represent promising novel biomarker candidates to predict functional cure 108 . However, more basic research is required to understand the biology of HBsAg isoforms and their clinical relevance, particularly as surrogate markers for HBsAg expression from cccDNA versus integrated HBV DNA and to ascertain if they provide additional diagnostic benefits for the staging of CHB or in monitoring response to current treatment modalities; they might prove to be valuable in monitoring future therapeutic approaches.
HBV biomarkers during treatment
Receiver operating characteristic curves showed that absolute HBV RNA levels were consistently superior to the change from baseline for predicting peg-IFNα response in patients 77 . No single biomarker seemed superior when comparing HBV RNA, HBV DNA, HBeAg and HBsAg. However, HBV RNA and HBsAg were more accurate at predicting non-responders than HBeAg and HBV DNA 77 . Furthermore, patients with CHB who were HBeAg negative and treated with peg-IFNα showed rapid HBV RNA decline that correlated with treatment response and long-term HBsAg loss 110 . This finding likely reflects the ability of peg-IFNα to act as an immune modulator and to lower HBV transcript levels. Similarly, HBcrAg was associated with treatment response for NUC, with and without peg-IFNα, in patients with CHB who were HBeAg positive and in those with CHB who were HBeAg negative 101 , 111 . However, HΒcrAg was not superior to HBsAg in predicting therapy response.
The different mechanisms of action of HBV drugs might affect the performance of HBV serum biomarkers, and this needs to be considered, particularly as new therapies targeting different aspects of the HBV life cycle are developed. For example, the reduction in RNA levels was consistently higher in patients treated with CAMs in combination with NUCs than in those treated with NUCs alone 112 . The reduction of serum HBV RNA during treatment with CAMs is consistent with their mechanism, which blocks pgRNA packaging into capsids as required for their secretion into blood 113 . In this case, serum RNA will no longer correlate with cccDNA levels or transcriptional activity but can serve to monitor target engagement.
Immunological serum biomarkers
In CHB, HBV persists with dynamic variations in hepatocellular injury with inflammation versus disease and/or virus control and the participation of multiple immune effectors and regulatory pathways 2 , 3 , 4 , 5 , 6 , 7 . Given the lack of safe and convenient access to the liver compartment, examining the serum immune markers is needed to gain mechanistic, clinical and prognostic insights.
As HBV is non-cytopathic, HBV-associated liver disease is largely immune mediated, with the host immune response being induced upon viral recognition (Fig. 3 ). HBV persistence is associated with global and virus-specific adaptive immune dysregulation or tolerance. In persistent HBV infection without a robust adaptive immune response, multiple inflammatory mechanisms can be activated to mediate hepatocellular injury 114 . The immune exhaustion, tolerance and pathogenic mechanisms with associated markers and cell subsets, summarized in Box 1 , also provide potential avenues for therapeutic immune restoration 115 .
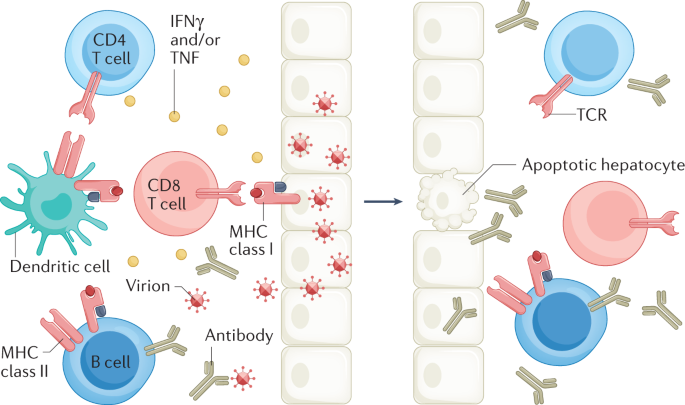
Control of hepatitis B virus (HBV) infection requires both cellular (CD4 + and CD8 + T cells) and humoral (antibody production by B cells) arms. Using both cytolytic and cytokine-mediated non-cytolytic mechanisms and major histocompatibility (MHC) class I and class II antigen recognition, CD8 + T cells have a primary effector role to kill and cure HBV-infected hepatocytes 7 , 114 . CD4 + T cells have a key regulatory role 144 , 183 . Neutralizing antibodies to hepatitis B surface antigen (anti-HBsAg) bind circulating virus, thereby reducing viral spread and providing protective immunity 184 . A key role for B cells in protective immunity to HBV has also been suggested by the high rate of HBV reactivation in patients undergoing B cell depletion with anti-CD20 (ref. 185 ). IFNγ, interferon-γ.
A challenge and opportunity is the measurement of biomarkers in serum that can reflect immunological activity in the liver. In theory, this could be achieved using cytokines, chemokines, and immune regulatory and metabolic factors that can be followed in relation to the clinical course of CHB and response to antiviral therapy (Box 2 ). Currently, none of these markers seems to be superior to other clinical and virological measures, and further investigations are required to evaluate the potential clinical role of these markers. However, they might collectively provide insights to host responses during novel HBV therapies with potential mechanistic and prognostic implications. In this regard, as a first option, serum biomarkers (for example, cytokines, chemokines, metabolic markers, soluble PD1, soluble CD14) might be more readily examined in clinical studies as only a small amount of blood is needed to measure hundreds of markers simultaneously in highly multiplexed assays 116 . A second option is the examination of cellular immune phenotype and function in peripheral blood. This approach can vary in complexity yet can be feasible at scale once optimized, and can range from a simple cytokine stimulation assay to assess HBV-specific T cells similar to what has been done for coronavirus disease 2019 (COVID-19) 117 , 118 , 119 or the diagnostic test for tuberculosis that uses an IFNγ release assay 120 , to a more complex phenotypic and functional analysis. Considering global and virus-specific immune dysfunction in CHB, a therapeutic goal is to achieve sustained virus control with immune restoration by suppressing viral antigen expression and the viral life cycle. Accordingly, there is a strong rationale to examine how the immune phenotype and function are affected by novel therapies, including immune-modulatory therapies such as Toll-like receptors agonists, therapeutic vaccines, checkpoint inhibitors and those targeting the viral life cycle with potential immune effect 121 . As a third option, despite growing challenges, direct sampling of the intrahepatic compartment (for example, through liver biopsy) is needed to visualize viral and immune markers simultaneously 122 , 123 , and such analyses can currently be done more comprehensively by using various emerging highly multiplexed and computational approaches 124 , 125 , 126 to better understand the spatial landscape of HBV and host responses in the liver.
Box 1 Mechanisms contributing to immune dysregulation or tolerance and leading to pathogenesis in CHB
Mechanisms of T cell dysfunction
Antigen-specific exhaustion with the induction of checkpoint molecules such as PD1 and CTLA4 (refs. 114 , 186 ) in addition to epigenetic changes.
T cell deletion through the pro-apoptotic protein Bcl2-interacting mediator (Bim) 187 or activated NK cells 188 .
Induction of regulatory T cells, cytokines and chemokines 189 , 190 , 191 .
Myeloid-derived suppressor cells that secrete soluble arginase 192 that can deplete arginine from T cells.
Mitochondrial alterations with further metabolic deficit 193 .
Mechanisms of B cell dysfunction or tolerance
Increased PD1 expression can lead to global and HBV-specific impairment of B cell differentiation and function 194 .
Excess HBsAg contributes to B cell exhaustion with HBeAg mediating antigen-specific immune tolerance 195 .
Regulatory and pathogenic mechanisms
Inflammatory cytokines and/or chemokines (for example, from dendritic cells and Kupffer cells) can increase hepatic inflammatory infiltrates, induce inflammatory NK cells and promote hepatocellular susceptibility to apoptosis.
Apoptosis can be induced through TRAILR2 (ref. 196 ), contributing to hepatocellular injury, with fluctuations in levels of ALT, HBV DNA, HBsAg and/or HBeAg, ultimately leading to liver disease progression.
NK cells can also kill HBV-specific T cells through NKG2D-dependent and TRAIL-dependent lysis 188 .
Damaged hepatocytes and myeloid-derived suppressor cells release soluble arginase, which depletes arginine and leads to suppression of T cell proliferation 114 , 191 , 192 , 197 , 198 .
Platelets can also promote accumulation of inflammatory cells in the liver, contributing to pathogenesis 199 .
Liver sinusoidal endothelial cells can prime CD4 + and CD8 + T cells with diverse effects, they can also produce IL-10 and express PDL1 with potential regulatory effect with activated immune cells expressing PD1 (refs. 200 , 201 ).
Altered phenotype and/or function of γδ T cells 202 and mucosal-associated invariant T cells 203 have been described in CHB, in association with clinical and/or therapeutic virus suppression.
Additional hepatic cells that participate in HBV immune pathogenesis include liver sinusoidal endothelial cells, myeloid-associated T cells and platelets, resulting in fibrogenesis.
ALT, alanine aminotransferase; CHB, chronic hepatitis B; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; NK, natural killer.
Box 2 Immunological serum markers to follow the natural history of HBV infection and outcome of antiviral therapy
Immunological markers to follow the natural history of HBV infection
IFNα, IL-8 and NK expression of TRAIL
Pathogenetic markers correlated with viral loads and flares of liver inflammation (ALT) 196 :
IFNα increased TRAIL expression in peripheral NK cells, which could induce apoptosis of hepatocytes expressing TRAIL-receptor.
Induction of TRAIL-receptor expression in HBV-infected liver, whereas is IL-8 shown to increase TRAIL-receptor expression in vitro.
Chemokines CXCL9–11 and IDO
Markers associated with hepatocellular injury, immune recruitment and potential antiviral activity 204 :
CXCL9–11 levels correlated with ALT and IDO activity.
CXCL10 positively correlated with increased levels of ALT and T cell expression of PD1 (refs. 205 , 206 ) as well as hepatic inflammatory score but negatively with serum HBV DNA and HBsAg 207 .
IDO is inducible in epithelial and plasmacytoid dendritic cells by IFNγ and/or TNF and has both regulatory and antiviral activities.
Metabolic markers (arginase and L-arginine)
Arginine depletion in the inflamed liver due to increased arginase as a potential mechanism for the global defect in CD8 T cell signalling and function in CHB:
L-Arginine is needed for T cell metabolism and survival.
Increased serum arginase activity and reduced serum L-arginine levels were associated with increased ALT activity and flares in patients with AHB and CHB 197 , 198 .
Increased arginase suppressed antiviral T cell function.
sPD1 and sPDL1
Soluble markers with a potential regulatory role in the PD1–PDL1 pathway 192 , 198 :
Binding of sPD1 to membrane-bound PDL1 on target tissues can block the regulatory interactions with PD1 expressed on activated immune cells or, alternatively, binding of sPDL1 to PD1-expressing immune cells can inhibit their interactions with membrane-bound PDL1 expressed by target tissues 208 .
Serum sPD1 levels were associated with persistently higher HBV load and higher HCC risk 209 .
Serum sPD1 levels were greater in CHB than in controls and positively correlated with levels of ALT, AST, total bilirubin, HBV DNA, AST to platelet ratio index ((AST/upper limit of the normal AST range) X 100/platelet count), fibrosis score Fib4, hepatic inflammatory score and fibrosis but negatively correlated with platelet count 210 .
As a caveat, there are assay-related issues that need to be resolved before clinical application, with additional head-to-head comparisons of the different immune assays needed to resolve discrepant sPD1 and sPDL1 levels observed in different studies 211 .
Immunological serum markers to follow the outcome of antiviral therapy
Greater serum levels at baseline and at the time of HBV DNA suppression in patients who achieved over 0.5 log decline in HBsAg on NUC therapy than those who did not 212 .
Decline in serum levels on antiviral therapy correlated with virological response to telbivudine treatment 205 .
Pre-treatment serum CXCL10 (also known as IP-10) levels were substantially greater in patients with CHB who achieved HBeAg clearance or HBsAg decline with peg-IFNα therapy 207 . Similarly, higher pre-treatment CXCL10 levels correlated with an increased probability of HBeAg loss after peg-IFNα therapy, with declines in HBV DNA, HBeAg and HBsAg being steeper in individuals with CXCL10 levels >150 pg/ml. However, this correlation only held for HBV infection without basal core promoter and/or precore mutants 213 .
Multivariate logistic regression analysis showed serum CXCL10 level to be an independent predictor of HBeAg clearance and HBsAg decline 207 .
Cytokines and chemokines
Substantial increases in CXCL13 and IL-21 levels were detected in patients with CHB who attained HBsAg seroconversion but not in patients with CHB with persistent HBsAg, including those with flares 214 .
Substantial increases in CD163, TNF, IL-12p70, IL-1α, IL-1β, IL-6, IL-18, IL-10, IL-2, IFNλ2, IFNα, FAS ligand, CXCL9, CXCL10, CXCL13 and CCL4 in patients with liver damage after stopping therapy 215 .
CXCL9, CXCL10, CXCL11, CXCL13 and IL-21 levels were elevated at the peak of AHB; IL-21 elevation was observed only in patients with self-limiting infection but not among those with chronic evolution.
Despite the small sample size, CXCL13 and IL-21 might be markers of functional cure for both AHB and CHB 214 .
Lower baseline sPD1 levels were associated with HBeAg clearance after 2 years of antiviral treatment in patients with HBeAg positive CHB 216 .
sCD14, a co-receptor for lipopolysaccharides, is a biomarker in infectious and inflammatory diseases that is produced by liver monocytes, macrophages and human hepatocytes.
sCD14 levels were substantially higher in AHB than in patients with CHB or healthy individuals as controls in one study 217 .
sCD14 level increased substantially at 12 weeks post-treatment compared to baseline in patients with CHB receiving peg-IFNα, with the fold change being substantially higher in responders than in non-responders.
sCD14 levels correlated with markers of hepatic inflammation and fibrosis in patients infected with HCV or HBV 218 .
AHB, acute hepatitis B; ALT, alanine aminotransferase; AST, aspartate aminotransferase; CHB, chronic hepatitis B; HBeAg, hepatitis B e antigen; HBsAg, hepatitis B surface antigen; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; IDO, indoleamine 2,3 oxygenase; IFNα, interferon-α; IFNγ, interferon-γ; NK, natural killer; NUC, nucleos(t)ide analogue; Peg-IFNα, pegylated IFNα; sCD14, secreted CD14; sPD1, soluble PD1; sPDL1, soluble PDL1.
Biomarkers of occult HBV infection
OBI is defined as the presence of cccDNA in the liver and/or HBV DNA in the blood of people who tested negative for HBsAg by currently available assays 33 , 127 . Statements on the biology and clinical effect of OBI suggested that the ideal diagnosis method for OBI is the detection of replication-competent HBV DNA in the liver 33 . The recommended methods included nested-PCR techniques to amplify at least three different viral genomic regions, real-time PCR assays or droplet digital PCR (ddPCR) assays. In each case, the assay must include primer sets enabling the detection of replication-competent HBV DNA 33 .
The diagnosis of OBI depends on the sensitivity of assays used to detect HBV DNA in liver tissue and/or blood samples and HBsAg in serum samples. Sensitivity for HBV DNA detection is improving with new technologies such as ddPCR assays. In a study of 100 transplant liver donors who were anti-HBc positive, OBI was diagnosed using four parallel nested PCRs to detect HBV surface, core, polymerase and X sequences 128 . Next, ddPCR was used to quantify cccDNA, which was detected in 52% (52 of 100) of the individuals who were OBI positive, with a median of 13 copies per 10 5 cells (95% CI 5–25) 128 . More sensitive HBsAg assays have also been developed. Commonly used assays detect HBsAg at 0.05 IU/ml; however, the newly developed assays can detect HBsAg with a sensitivity of 0.005 IU/ml (ref. 129 ). These more sensitive assays can improve the detection rate of low-level HBsAg, HBsAg variants, and HBsAg with anti-HBs 130 , 131 and can provide improved detection of OBI 132 .
In addition to HBsAg assays, inadequately sensitive HBV DNA assays can lead to false-negative HBV DNA results and a missed OBI diagnosis 30 . Commercially available real-time PCR-based assays for serum HBV DNA detection are sufficiently sensitive to detect many (but not all) OBI cases. ddPCR assays might increase the rate of OBI detection and need to be evaluated systematically in the OBI setting. In addition, consideration should be given to the need to diagnose OBI by re-screening samples by ddPCR, which is only accessible via research facilities but not routinely available in diagnostic laboratories 31 , 133 .
Lessons can be learned from HBV reactivation studies in patients with chronic hepatitis C treated with direct-acting antiviral agents. Nested-PCR testing of longitudinally collected serum samples from 40 patients revealed that serum HBV DNA was intermittently detected in 25% (10 of 40) at baseline and 52.5% (21 of 40) at 3 months after termination of direct-acting antiviral treatment. Moreover, HBsAg and HBcrAg biomarkers were negative at baseline and remained persistently negative (without any fluctuation) in all the serially collected serum samples 134 . Thus, as HBV DNA is usually present in low concentrations and can only be intermittently detected in people with OBI, testing blood samples collected at more than one timepoint and testing DNA extracts from at least 1 ml of serum or plasma is recommended for OBI diagnosis 33 , 127 . Indeed, it should be considered that OBI is characterized by periods of transient HBV viraemia alternating with periods in which the viral DNA is undetectable in blood 135 , 136 , 137 , 138 . Moreover, evidence demonstrated an association between the reappearance of circulating HBV DNA and phases of ALT serum level increases, suggesting a role in the transient reactivation of HBV replication in liver cell injury 136 , 137 . Furthermore, Candotti et al. described nine cases of undetected HBV transfusion-transmission from OBI-positive blood donation despite the use of highly sensitive HBsAg and HBV DNA screening assays 30 . Importantly, the availability of archive samples from both donors and recipients and large-volume (2–24 ml) follow-up donor samples enabled the researchers to detect HBV transfusion-transmitted infection associated with extremely low HBV DNA loads. These results led the researchers to conclude that, until more sensitive assays become available, long-term archiving of large-volume pretransfusion plasma samples from both donors and recipients is essential to identify transfusion-transmission of undetected OBI to limit delays in the therapeutic management of patients with HBV infection 30 .
As molecular tests are not always available, there is a strong consensus that detection of anti-HBc in the blood can be used as a surrogate biomarker for OBI in blood and/or organ donors and in people initiating immunosuppressive therapies 33 , 139 . Although serum levels of anti-HBc correlate with cccDNA positivity 128 , the absence of anti-HBc does not rule out OBI 33 . High baseline levels of anti-HBc (and low or absent anti-HBs) were shown to predict HBV reactivation in 36 of 192 patients with lymphoma and resolved HBV infection receiving B cell-depleting chemotherapies (hazard ratio of 17.29 for HBV reactivation (95% CI 3.92–76.30; P <0.001)) 140 , 141 . Anti-HBc quantification and analysis of circulating HBV-specific T cells in patients who are HBsAg negative might be interesting biomarkers but require further confirmation in the setting of OBI. Currently, HBV DNA is the only reliable diagnostic marker of OBI and a standardized diagnostic procedure for OBI remains an important unmet clinical need.
Biomarkers of liver cancer
Chronic HBV infection causes liver cancer, the sixth most common cancer and the third leading cause of cancer-related death worldwide. HCC is more common in men (2–4 times higher incidence than in women) and the prognosis is poor in all regions of the world, with incidence and mortality rates being roughly equivalent. The median survival of patients with early HCC is >5 years but is <1 year when detected at an advanced stage. Due to a lack of appropriate biomarkers, most HCC cases are detected at late stages and not when the tumour is still localized and treatment options are more effective 142 .
Currently, HCC surveillance relies on a limited armoury of serum biomarkers and/or imaging of the affected liver. Cancer biomarkers are detected in the blood, urine or other body fluids and can indicate the presence of cancer or predict the risk of cancer development 143 . Ideally, biomarkers should enable early detection of cancer by screening healthy or high-risk populations, confirming the diagnosis or identifying a specific type of cancer, predicting prognosis, monitoring treatment response, and detecting early recurrence 142 .
The identification of biomarkers for the early detection of cancer requires the following steps 131 :
Phase 1: Preclinical exploratory studies: to identify promising biomarker candidates.
Phase 2: Clinical assay and validation: to detect the disease versus controls (for example, distinguish HCC from non-HCC).
Phase 3: Retrospective longitudinal repository studies: to detect preclinical disease by retrospective analysis.
Phase 4: Prospective screening studies: to determine the detection rate of the assay (sensitivity and specificity).
Phase 5: Cancer control studies: to assess the effect of screening on reducing the disease burden in the target population.
Over the past several decades, AFP has been the most extensively studied and most commonly used HCC biomarker. It has been utilized for the assessment risk of HCC in patients with cirrhosis, as a screening tool for the early detection of HCC, and as a diagnostic and prognostic tool for HCC 144 . In addition to AFP, a number of novel biomarkers for HCC diagnosis and monitoring are in different phases of development. Serum biomarkers currently in phase 2 (clinical assay and validation) include osteopontin, midikine, dikkopf 1, glypican 3, α1 fucosidase, Golgi protein 73 and squamous cell carcinoma antigen 145 , 146 , 147 . Serum biomarkers at more advanced stages of development include AFP (phase 5), Lens culinaris agglutinin fraction of AFP (AFP-L3) (phase 2/3) and des-γ-carboxy prothrombin (DCP) (phase 2/3) 142 . Genetic and cellular biomarkers (so-called liquid biopsy) under investigation include circulating tumour cells, circulating tumour DNA, microRNA and long non-coding RNA 148 .
AFP is the best characterized and most widely used serum biomarker for HCC surveillance. However, its effectiveness is limited as not all HCCs secrete AFP 149 . In addition, AFP serum levels can be elevated in patients with chronic hepatitis or cirrhosis. However, with the advent of highly effective NUCs for the treatment of CHB, elevated on-treatment AFP levels were shown in a large retrospective-prospective study to be a specific marker for HCC because falsely elevated AFP levels in 1,531 patients receiving entecavir were minimized compared to 424 patients that received no treatment, suggesting that, in this group of patients, a lower AFP cut off value could be used 150 . Elevated on-treatment AFP is a specific tumour marker for HCC in patients with CHB receiving entecavir 150 . There is little debate that AFP should not be used alone in HCC surveillance, but it has been debated whether AFP should be included in HCC surveillance due to its suboptimal sensitivity (39–65%) and specificity (76–97%) 151 . However, most studies show a benefit of combining AFP with ultrasonography 152 . Various factors can influence the performance of AFP as an HCC biomarker, including patient demographics, aetiology of underlying liver disease, severity of liver disease (cirrhosis, chronic hepatitis, serum ALT values), antiviral therapy, and tumour stage and biology 153 . In turn, according to the size of the tumour, the sensitivity of ultrasonography imaging for detecting HCC at an early stage is highly variable, ranging from 21% to 89% across studies included in a meta-analysis published in 2018 (ref. 152 ). It is largely operator dependent, based on the skill of the sonographer and influenced by patient characteristics, including obesity, liver nodularity and presence of ascites 154 , 155 , 156 . A meta-analysis of 32 studies comprising 13,367 patients collected worldwide showed that ultrasonography alone detected early-stage HCC with a sensitivity of 45% compared to 63% when ultrasonography was combined with AFP ( P = 0.002) 152 . The improved sensitivity was associated with a decrease in specificity (84% versus 92%). However, the addition of AFP to ultrasonography significantly increased the sensitivity of early HCC detection, suggesting this might be the preferred surveillance strategy for patients with cirrhosis 152 . Other factors to consider are the value of single timepoint versus longitudinal analysis and tailoring cut-offs according to liver disease aetiology, severity and antiviral therapy. The diagnostic value of AFP for detecting HCC was also improved when used in combination with the level of serum protein induced by vitamin K absence or antagonist II (PIVKAII) in patients of European descent 157 and Asian 158 (South Korea) patients with cirrhosis.
Longitudinal determinations also improve AFP performance as an HCC biomarker 159 . A phase III biomarker study evaluating AFP, AFP-L3, DCP and their combinations for the early detection of HCC in prospectively collected longitudinal samples from 689 patients with cirrhosis or CHB 160 showed that a combination of AFP and AFP-L3 at diagnosis differentiated early-stage HCC from cirrhosis better than each biomarker individually. Investigating the sensitivity and specificity of ultrasonography alone or in combination with biomarkers showed that adding AFP to ultrasonography increased the sensitivity to 88.6%, and adding AFP plus AFP-L3 to ultrasonography increased the sensitivity to 94.3% 160 .
In summary, although the addition of AFP to ultrasonography imaging markedly improved the early detection of HCC, results are still suboptimal and new biomarkers to predict early-onset HCC are required. Longitudinal determination of AFP increases the sensitivity and specificity for HCC surveillance but optimal cut-offs for AFP and other biomarkers of HCC surveillance in patients with suppressed HBV and minimal hepatic inflammation are unclear. Given the high degree of heterogeneity of HCC, the combination of AFP with other biomarkers and clinical parameters improves the sensitivity and specificity of surveillance for early HCC detection.
The road forward
To map a way forward, the clinical utility of both classic and emerging viral and immunological biomarkers of HBV infection, with respect to the course of infection, disease progression, and response to current and emerging treatments, was appraised in two-panel discussions. The panels discussed the latest advances, knowledge gaps, and key challenges and opportunities for improvement were identified by addressing three key questions: do we have the appropriate biomarkers to measure HBV cccDNA, and are the emerging biomarkers relevant for measuring the mechanism of action of new drugs? What are the key biomarkers that require further research to have the strongest clinical effect? Finally, how urgent is the need for predictive immunological biomarkers for inflammation or antiviral response? The strengths and weakness of current biomarkers for addressing each of these questions are presented in Table 2 , with a Roadmap outlining the actions required to address unmet clinical needs presented in Table 3 .
For patients with CHB undergoing, or ceasing current NUC therapy, no single biomarker is currently clearly superior in predicting treatment response or relapse across all stages of CHB. There is global consensus that achieving an HBV cure will require combination therapies, targeting different steps of the HBV replication cycle, and stimulating host immune responses to neutralize HBV infection and/or safely eradicate HBV-infected cells 26 . In turn, approaches that affect viral and immunological targets will require a combination of viral and immunological biomarkers to monitor progress towards the new treatment end points.
For the immediate future, HBeAg, HBsAg and HBV DNA will remain the most important viral serum biomarkers used in natural history and treatment end points because they are best validated to reflect outcomes. It is evident that biomarkers have different importance in various disease stages (HBeAg positive versus HBeAg negative), HBV genotypes and for different treatment modalities. On the other hand, HBV RNA and HBcrAg are expected to correlate with treatment response to current and new therapies but might not outperform HBsAg in predicting treatment outcome when each is taken in isolation. HBV RNA, HBcrAg and/or HBcAg tests need to be validated and standardized, and their sensitivity optimized. New biomarkers can help to dissect the mechanism of action of new drugs. Another possible benefit of these emerging biomarkers is their clinical usefulness in combination with conventional markers such as the demonstrated superiority of combining pgRNA and HBsAg levels at the time of stopping NUC for the prediction of off-treatment viral relapse 71 .
It is also important that biomarkers are validated for all major HBV genotypes, which exhibit marked differences in HBV natural history, disease progression and treatment response to peg-IFNα therapy 161 . Until new immune-mediated therapies are developed, peg-IFNα is likely to have a place in combination treatment regimens, yet it is only effective in patients with HBV infection of genotypes A and B and least effective for genotypes C and D. Although HBV sequencing and genotyping are not currently routinely undertaken prior to treatment initiation, this might need to be considered if peg-IFNα is included in treatment combinations. Although not routinely available in all settings, next-generation sequencing of complete HBV genomes prior to the initiation of antiviral therapy also shows promise as a biomarker of treatment response on NUCs, with the identification of basal core promoter mutations even at a low frequency associated with reduced likelihood of functional cure 162 and ALT flare 163 on therapy. As new curative therapies for HBV are developed, the effect of HBV variants on treatment response warrants further investigation where access to next-generation sequencing technology is available.
Although a number of biomarkers have been described that can monitor the course of HBV infection, it is vital, when interpreting their kinetics and variations, to determine whether they are informing on target engagement of the therapeutic agent and/or are a reflection of intrahepatic replication and immune control. The emerging biomarkers will have an important role as exploratory markers for end points and mode-of-action studies. No one biomarker yet fits all novel antiviral modalities, and an integrative approach might be necessary because the serological marker used is dependent on the mode of action of the antiviral drugs. More translational studies are required, and the relevance of these assays in the various phases of CHB natural history and in individuals of different ethnicities, age groups, sex and HBV genotypes is yet to be determined.
Currently, there are no serum or liver immunological biomarkers that are superior to clinical and virological markers in following the natural history of CHB and in monitoring therapy and HBV-specific immune responses. Markers that reflect the liver compartment will become increasingly important as access to liver tissue and standard liver biopsy become more difficult, with fine-needle aspiration (also known as fine-needle biopsy) showing promise. Monitoring intrahepatic activity will become increasingly important as therapies targeting HBV cccDNA are developed. However, in the interim, consideration should also be given to identifying the most appropriate biomarkers for treatment response using finite therapies that might reduce HBV DNA to below the limit of quantification but where HBsAg remains detectable, currently defined as a ‘partial functional cure’ 164 . ‘Partial functional cure’ might represent an important interim step as we progress on the path to increased rates of functional cure with new finite therapies. It is likely that pgRNA, HBcrAg and its HBcAg component as emerging biomarkers as well as additional yet-unidentified markers will have an important role; therefore, an HBcAg-specific biomarker would be a valuable additional tool to help de-convolute the multiple components of the current HBcrAg assay. Biomarkers of OBI are also required as are markers that will predict the likelihood of progression to HCC, enabling earlier interventions and more accurate risk assessment. Finally, all these resources need to be made available to all persons living with CHB in an equitable and fair manner, and particularly in resource-limited settings, where much of the burden of HBV worldwide lies, with a pressing need for POC biomarkers. In partnership with the HBV-affected community, academia, clinicians, the pharmaceutical and biotech industries, and organizations such as the HBV Forum and the Hepatitis B Foundation , ICE-HBV will work with all stakeholders to ensure this indeed occurs.
Tong, S. & Revill, P. Overview of hepatitis B viral replication and genetic variability. J. Hepatol. 64 , S4–S16 (2016).
Article CAS PubMed PubMed Central Google Scholar
Faure-Dupuy, S., Lucifora, J. & Durantel, D. Interplay between the hepatitis B virus and innate immunity: from an understanding to the development of therapeutic concepts. Viruses https://doi.org/10.3390/v9050095 (2017).
Article PubMed PubMed Central Google Scholar
Bertoletti, A. & Ferrari, C. Adaptive immunity in HBV infection. J. Hepatol. 64 , S71–S83 (2016).
Article CAS PubMed Google Scholar
Maini, M. K. & Gehring, A. J. The role of innate immunity in the immunopathology and treatment of HBV infection. J. Hepatol. 64 , S60–S70 (2016).
Kuipery, A., Gehring, A. J. & Isogawa, M. Mechanisms of HBV immune evasion. Antivir. Res. 179 , 104816 (2020).
Rehermann, B. & Thimme, R. Insights from antiviral therapy into immune responses to hepatitis B and C virus infection. Gastroenterology 156 , 369–383 (2019).
Bengsch, B. & Chang, K. M. Evolution in our understanding of hepatitis B virus virology and immunology. Clin. Liver Dis. 20 , 629–644 (2016).
Article PubMed Google Scholar
Grossi, G., Vigano, M., Loglio, A. & Lampertico, P. Hepatitis B virus long-term impact of antiviral therapy nucleot(s)ide analogues (NUCs). Liver Int. 37 (Suppl. 1), 45–51 (2017).
Lucifora, J. et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343 , 1221–1228 (2014).
Lok, A. S., Zoulim, F., Dusheiko, G. & Ghany, M. G. Hepatitis B cure: from discovery to regulatory approval. J. Hepatol. 67 , 847–861 (2017).
Revill, P. A. et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol. Hepatol. 4 , 545–558 (2019).
WHO. Interim Guidance for Country Validation of Viral Hepatitis Elimination https://www.who.int/publications/i/item/9789240028395 (2021).
Thomas, D. L. Global elimination of chronic hepatitis. N. Engl. J. Med. 380 , 2041–2050 (2019).
Revill, P. A., Penicaud, C., Brechot, C. & Zoulim, F. Meeting the challenge of eliminating chronic hepatitis B infection. Genes https://doi.org/10.3390/genes10040260 (2019).
Hepatitis B Foundation. Hepatitis B Foundation Drugwatch, https://www.hepb.org/treatment-and-management/drug-watch/ (2022).
Testoni, B., Levrero, M. & Zoulim, F. Challenges to a cure for HBV infection. Semin. Liver Dis. 37 , 231–242 (2017).
Sommer, G. & Heise, T. Posttranscriptional control of HBV gene expression. Front. Biosci. 13 , 5533–5547 (2008).
Yuen, M. F. et al. Hepatitis B virus infection. Nat. Rev. Dis. Prim. 4 , 18035 (2018).
Wooddell, C. I. et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aan0241 (2017).
Gish, R. G. et al. Chronic hepatitis B: virology, natural history, current management and a glimpse at future opportunities. Antivir. Res. 121 , 47–58 (2015).
Liu, D. et al. Clinical relevance of the in situ assay for HBV DNA: a cross-sectional study in patients with chronic hepatitis B. J. Clin. Pathol. 73 , 813–818 (2020).
Zhang, X. et al. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J. Clin. Investig. 126 , 1079–1092 (2016).
Bowden, S., Jackson, K., Littlejohn, M. & Locarnini, S. Quantification of HBV covalently closed circular DNA from liver tissue by real-time PCR. Methods Mol. Med. 95 , 41–50 (2004).
CAS PubMed Google Scholar
Werle-Lapostolle, B. et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology 126 , 1750–1758 (2004).
Coffin, C. S., Zhou, K. & Terrault, N. A. New and old biomarkers for diagnosis and management of chronic hepatitis B virus infection. Gastroenterology 156 , 355–368 e353 (2019).
Xu, H. et al. Role of anti-HBs in functional cure of HBeAg+chronic hepatitis B patients infected with HBV genotype A. J. Hepatol. 76 , 34–45 (2022).
Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 69 , 89–95 (2001).
Article Google Scholar
Hong, X. et al. Characterization of hepatitis B precore/core-related antigens. J. Virol. https://doi.org/10.1128/JVI.01695-20 (2021).
Candotti, D., Assennato, S. M., Laperche, S., Allain, J. P. & Levicnik-Stezinar, S. Multiple HBV transfusion transmissions from undetected occult infections: revising the minimal infectious dose. Gut 68 , 313–321 (2019).
Piermatteo, L. et al. Droplet digital PCR assay as an innovative and promising highly sensitive assay to unveil residual and cryptic HBV replication in peripheral compartment. Methods 201 , 74–81 (2022).
Carey, I. et al. Pregenomic HBV RNA and hepatitis B core-related antigen predict outcomes in hepatitis B e antigen-negative chronic hepatitis B patients suppressed on nucleos(t)ide analogue therapy. Hepatology 72 , 42–57 (2020).
Raimondo, G. et al. Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J. Hepatol. 71 , 397–408 (2019).
WHO. Prevention of Mother-to-Child Transmission of Hepatitis B Virus: Guidelines on Antiviral Prophylaxis in Pregnancy https://apps.who.int/iris/bitstream/handle/10665/333391/9789240002708-eng.pdf?sequence=1&isAllowed=y (2020).
Cornberg, M. et al. The role of quantitative hepatitis B surface antigen revisited. J. Hepatol. 66 , 398–411 (2017).
WHO. WHO Guidelines on Hepatitis B and C Testing https://apps.who.int/iris/bitstream/handle/10665/254621/9789241549981-eng.pdf?sequence=1 (2017).
Kramvis, A. Challenges for hepatitis B virus cure in resource-limited settings in sub-Saharan Africa. Curr. Opin. Hiv. AIDS 15 , 185–192 (2020).
Kosack, C. S., Page, A. L. & Klatser, P. R. A guide to aid the selection of diagnostic tests. Bull. World Health Organ. 95 , 639–645 (2017).
Sonderup, M. W. & Spearman, C. W. Global disparities in hepatitis B elimination — a focus on Africa. Viruses https://doi.org/10.3390/v14010082 (2022).
Scheiblauer, H. et al. Performance evaluation of 70 hepatitis B virus (HBV) surface antigen (HBsAg) assays from around the world by a geographically diverse panel with an array of HBV genotypes and HBsAg subtypes. Vox Sang. 98 , 403–414 (2010).
Chevaliez, S. & Pawlotsky, J. M. New virological tools for screening, diagnosis and monitoring of hepatitis B and C in resource-limited settings. J. Hepatol. 69 , 916–926 (2018).
Alavian, S. M., Carman, W. F. & Jazayeri, S. M. HBsAg variants: diagnostic-escape and diagnostic dilemma. J. Clin. Virol. 57 , 201–208 (2013).
Thibault, V., Servant-Delmas, A., Ly, T. D., Roque-Afonso, A. M. & Laperche, S. Performance of HBsAg quantification assays for detection of Hepatitis B virus genotypes and diagnostic escape-variants in clinical samples. J. Clin. Virol. 89 , 14–21 (2017).
Lange, B. et al. Diagnostic accuracy of detection and quantification of HBV-DNA and HCV-RNA using dried blood spot (DBS) samples - a systematic review and meta-analysis. BMC Infect. Dis. 17 , 693 (2017).
Shimakawa, Y. et al. Analytical validation of hepatitis B core-related antigen (HBcrAg) using dried blood spots (DBS). J. Viral Hepat. 28 , 837–843 (2021).
Jackson, K., Tekoaua, R., Li, X. & Locarnini, S. Real-world application of the Xpert(R) HBV viral load assay on serum and dried blood spots. J. Med. Virol. 93 , 3707–3713 (2021).
Abravanel, F. et al. Performance of the Xpert HBV Viral Load assay versus the Aptima Quant assay for quantifying hepatitis B virus DNA. Diagn. Microbiol. Infect. Dis. 96 , 114946 (2020).
Zhu, X. et al. Prospective evaluation of FibroScan for the diagnosis of hepatic fibrosis compared with liver biopsy/AST platelet ratio index and FIB-4 in patients with chronic HBV infection. Dig. Dis. Sci. 56 , 2742–2749 (2011).
Gerlich, W. H., Glebe, D., Kramvis, A. & Magnius, L. O. Peculiarities in the designations of hepatitis B virus genes, their products, and their antigenic specificities: a potential source of misunderstandings. Virus Genes 56 , 109–119 (2020).
Seeger, C. & Mason, W. S. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64 , 51–68 (2000).
Wang, J. et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J. Hepatol. 65 , 700–710 (2016).
Jansen, L. et al. Hepatitis B virus pregenomic RNA is present in virions in plasma and is associated with a response to pegylated interferon Alfa-2a and nucleos(t)ide analogues. J. Infect. Dis. 213 , 224–232 (2016).
Prakash, K. et al. High serum levels of pregenomic RNA reflect frequently failing reverse transcription in hepatitis B virus particles. Virol. J. 15 , 86 (2018).
Lam, A. M. et al. Hepatitis B virus capsid assembly modulators, but not nucleoside analogs, inhibit the production of extracellular pregenomic RNA and spliced RNA variants. Antimicrob. Agents Chemother. https://doi.org/10.1128/AAC.00680-17 (2017).
Stadelmayer, B. et al. Full-length 5’RACE identifies all major HBV transcripts in HBV-infected hepatocytes and patient serum. J. Hepatol. 73 , 40–51 (2020).
Wang, J. et al. Relationship between serum HBV-RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. J. Hepatol. 68 , 16–24 (2018).
Article CAS Google Scholar
Hacker, H. J., Zhang, W., Tokus, M., Bock, T. & Schroder, C. H. Patterns of circulating hepatitis B virus serum nucleic acids during lamivudine therapy. Ann. N. Y. Acad. Sci. 1022 , 271–281 (2004).
Niu, C. et al. The Smc5/6 complex restricts HBV when localized to ND10 without inducing an innate immune response and is counteracted by the HBV X protein shortly after infection. PLoS One 12 , e0169648 (2017).
Wang, J. et al. HBV RNA virion-like particles produced under nucleos(t)ide analogues treatment are mainly replication-deficient. J. Hepatol. 68 , 847–849 (2018).
Gunther, S., Sommer, G., Iwanska, A. & Will, H. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology 238 , 363–371 (1997).
Lim, C. S. et al. Quantitative analysis of the splice variants expressed by the major hepatitis B virus genotypes. Microb. Genom. https://doi.org/10.1099/mgen.0.000492 (2021).
Bai, L. et al. Extracellular hepatitis B virus RNAs are heterogeneous in length and circulate as capsid-antibody complexes in addition to virions in chronic hepatitis B patients. J. Virol. https://doi.org/10.1128/JVI.00798-18 (2018).
Butler, E. K. et al. Hepatitis B virus serum DNA and RNA levels in nucleos(t)ide analog-treated or untreated patients during chronic and acute infection. Hepatology 68 , 2106–2117 (2018).
van Bommel, F. et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology 61 , 66–76 (2015).
Mak, L. Y. et al. HBV RNA profiles in patients with chronic hepatitis B under different disease phases and antiviral therapy. Hepatology 73 , 2167–2179 (2021).
van Campenhout, M. J. H. et al. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology 68 , 839–847 (2018).
Anderson, M. et al. Circulating pregenomic HBV RNA is primarily full-length in chronic hepatitis B patients undergoing nucleos(t)ide analogue therapy. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa1015 (2020).
Wang, J. et al. Natural history of serum HBV-RNA in chronic HBV infection. J. Viral Hepat. 25 , 1038–1047 (2018).
Volz, T. et al. Impaired intrahepatic hepatitis B virus productivity contributes to low viremia in most HBeAg-negative patients. Gastroenterology 133 , 843–852 (2007).
Cathcart, A. L. et al. Evaluation of serum HBV RNA and HBcrAg in chronic hepatitis B patients achieving different serological outcomes on tenofovir disoproxil fumarate (TDF). J. Hepatol . 66 , S476 (2017).
Seto, W. K. et al. Role of serum HBV RNA and hepatitis B surface antigen levels in identifying Asian patients with chronic hepatitis B suitable for entecavir cessation. Gut 70 , 775–783 (2021).
van Bömmel, F. et al. HBV RNA can still be quantified in serum in HBeAg negative patients after suppression of HBV DNA by nuleos(t)ide analogues for up to 10 years. Hepatology 68 (Suppl.), 273A (2018).
Google Scholar
Fan, R. et al. Association between negative results from tests for hBV DNA and RNA and durability of response after discontinuation of nucles(t)ide analogue therapy. Clin. Gastroenterol. Hepatol. 18 , 719–727.e7 (2020).
Klumpp, K. et al. Efficacy of NVR 3-778, alone and in combination with pegylated interferon, vs entecavir in uPA/SCID mice with humanized livers and HBV infection. Gastroenterology 154 , 652–662.e8 (2018).
Yuen, M. F. et al. Antiviral activity, safety, and pharmacokinetics of capsid assembly modulator NVR 3-778 in patients with chronic HBV infection. Gastroenterology 156 , 1392–1403.e7 (2019).
Giersch, K., Allweiss, L., Volz, T., Dandri, M. & Lutgehetmann, M. Serum HBV pgRNA as a clinical marker for cccDNA activity. J. Hepatol. 66 , 460–462 (2017).
van Bommel, F. et al. Serum HBV RNA as a predictor of peginterferon Alfa-2a response in patients with HBeAg-positive chronic hepatitis B. J. Infect. Dis. 218 , 1066–1074 (2018).
Hu, J. & Liu, K. Complete and incomplete hepatitis B virus particles: formation, function, and application. Viruses https://doi.org/10.3390/v9030056 (2017).
Liu, Y. Y. & Liang, X. S. Progression and status of antiviral monitoring in patients with chronic hepatitis B: from HBsAg to HBV RNA. World J. Hepatol. 10 , 603–611 (2018).
Suzuki, F., Miyakoshi, H., Kobayashi, M. & Kumada, H. Correlation between serum hepatitis B virus core-related antigen and intrahepatic covalently closed circular DNA in chronic hepatitis B patients. J. Med. Virol. 81 , 27–33 (2009).
Hige, S. et al. Sensitive assay for quantification of hepatitis B virus mutants by use of a minor groove binder probe and peptide nucleic acids. J. Clin. Microbiol. 48 , 4487–4494 (2010).
Testoni, B. et al. Serum hepatitis B core-related antigen (HBcrAg) correlates with covalently closed circular DNA transcriptional activity in chronic hepatitis B patients. J. Hepatol. 70 , 615–625 (2019).
Mak, L. Y. & Yuen, M. F. Letter: serum HBcrAg is a useful marker for disease monitoring, predicting treatment response and disease outcome of chronic hepatitis B virus infection-authors’ reply. Aliment. Pharmacol. Ther. 47 , 1720–1721 (2018).
Mak, L. Y. et al. Review article: hepatitis B core-related antigen (HBcrAg): an emerging marker for chronic hepatitis B virus infection. Aliment. Pharmacol. Therapeut. 47 , 43–54 (2018).
Maasoumy, B. et al. Hepatitis B core-related antigen (HBcrAg) levels in the natural history of hepatitis B virus infection in a large European cohort predominantly infected with genotypes A and D. Clin. Microbiol. Infect. 21 , 606.e1-10 (2015).
Wong, G. L., Wong, V. W. & Chan, H. L. Virus and host testing to manage chronic hepatitis B. Clin. Infect. Dis. 62 (Suppl. 4), S298–305 (2016).
Chen, E. Q. et al. Serum hepatitis B core-related antigen is a satisfactory surrogate marker of intrahepatic covalently closed circular DNA in chronic hepatitis B. Sci. Rep. 7 , 173 (2017).
Suzuki, Y. et al. Hepatitis B virus (HBV)-infected patients with low hepatitis B surface antigen and high hepatitis B core-related antigen titers have a high risk of HBV-related hepatocellular carcinoma. Hepatol. Res. 49 , 51–63 (2019).
Seto, W. K. et al. Linearized hepatitis B surface antigen and hepatitis B core-related antigen in the natural history of chronic hepatitis B. Clin. Microbiol. Infect. 20 , 1173–1180 (2014).
Riveiro-Barciela, M. et al. Serum hepatitis B core-related antigen is more accurate than hepatitis B surface antigen to identify inactive carriers, regardless of hepatitis B virus genotype. Clin. Microbiol. Infect. 23 , 860–867 (2017).
Brunetto, M. R. et al. Incremental value of HBcrAg to classify 1582 HBeAg-negative individuals in chronic infection without liver disease or hepatitis. Aliment. Pharmacol. Therapeut. 53 , 733–744 (2021).
CAS Google Scholar
Chuaypen, N. et al. Predictive role of serum HBsAg and HBcrAg kinetics in patients with HBeAg-negative chronic hepatitis B receiving pegylated interferon-based therapy. Clin. Microbiol. Infect. 24 , 306.e7–306.e13 (2018).
Inoue, T. et al. Clinical efficacy of a novel, high-sensitivity HBcrAg assay in the management of chronic hepatitis B and HBV reactivation. J. Hepatol. https://doi.org/10.1016/j.jhep.2021.02.017 (2021).
Honda, M. et al. Hepatitis B virus (HBV) core-related antigen during nucleos(t)ide analog therapy is related to intra-hepatic HBV replication and development of hepatocellular carcinoma. J. Infect. Dis. 213 , 1096–1106 (2016).
Tseng, T. C. et al. Serum hepatitis B core-related antigen level stratifies risk of disease progression in chronic hepatitis B patients with intermediate viral load. Aliment. Pharmacol. Therapeut. 53 , 908–918 (2021).
Tada, T. et al. Hepatitis B virus core-related antigen levels predict progression to liver cirrhosis in hepatitis B carriers. J. Gastroenterol. Hepatol. 33 , 918–925 (2018).
Hosaka, T. et al. Impact of hepatitis B core-related antigen on the incidence of hepatocellular carcinoma in patients treated with nucleos(t)ide analogues. Aliment. Pharmacol. Therapeut. 49 , 457–471 (2019).
Tseng, T. C. et al. High level of hepatitis B core-related antigen associated with increased risk of hepatocellular carcinoma in patients with chronic HBV infection of intermediate viral load. Gastroenterology 157 , 1518–1529.e3 (2019).
Hosaka, T. Letter: impact of hepatitis B core-related antigen on the incidence of hepatocellular carcinoma in patients treated with nucleos(t)ide analogues-further clarifications needed. Authors’ reply. Aliment. Pharmacol. Therapeut. 50 , 233 (2019).
Wong, D. K. et al. Hepatitis B virus core-related antigen as a surrogate marker for covalently closed circular DNA. Liver Int. 37 , 995–1001 (2017).
van Campenhout, M. J. et al. Hepatitis B core-related antigen levels are associated with response to entecavir and peginterferon add-on therapy in hepatitis B e antigen-positive chronic hepatitis B patients. Clin. Microbiol. Infect. 22 , 571.e5-9 (2016).
PubMed Google Scholar
Matsuzaki, T. et al. Significance of hepatitis B virus core-related antigen and covalently closed circular DNA levels as markers of hepatitis B virus re-infection after liver transplantation. J. Gastroenterol. Hepatol. 28 , 1217–1222 (2013).
Kimura, T. et al. Hepatitis B virus DNA-negative dane particles lack core protein but contain a 22-kDa precore protein without C-terminal arginine-rich domain. J. Biol. Chem. 280 , 21713–21719 (2005).
Kimura, T. et al. Sensitive enzyme immunoassay for hepatitis B virus core-related antigens and their correlation to virus load. J. Clin. Microbiol. 40 , 439–445 (2002).
Fanning, G. C., Zoulim, F., Hou, J. & Bertoletti, A. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat. Rev. Drug Discov. 18 , 827–844 (2019).
Hong, X. et al. Characterization and application of precore/core-related antigens in animal models of hepatitis B virus infection. Hepatology https://doi.org/10.1002/hep.31720 (2021).
Pfefferkorn, M. et al. Quantification of large and middle proteins of hepatitis B virus surface antigen (HBsAg) as a novel tool for the identification of inactive HBV carriers. Gut 67 , 2045–2053 (2018).
Pfefferkorn, M. et al. Composition of HBsAg is predictive of HBsAg loss during treatment in patients with HBeAg-positive chronic hepatitis B. J. Hepatol. 74 , 283–292 (2021).
Hassemer, M. et al. Comparative characterization of hepatitis B virus surface antigen derived from different hepatitis B virus genotypes. Virology 502 , 1–12 (2017).
Farag, M. S. et al. Hepatitis B virus RNA as early predictor for response to PEGylated interferon Alfa in HBeAg negative chronic hepatitis B. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciaa013 (2020).
van Campenhout, M. J. H. et al. Hepatitis B core-related antigen monitoring during peginterferon alfa treatment for HBeAg-negative chronic hepatitis B. J. Viral Hepat. 26 , 1156–1163 (2019).
Zhang, M. Efficacy and safety of GLS4/ritonavir combined with entecavir in HBeAg-positive patients with chronic hepatitis B: interim results from phase 2b, multi-center study. J. Hepatol. 73 , s878 (2020).
Taverniti, V. et al. Capsid assembly modulators as antiviral agents against HBV: molecular mechanisms and clinical perspectives. J. Clin. Med. https://doi.org/10.3390/jcm11051349 (2022).
Ghany, M. G. et al. Serum alanine aminotransferase flares in chronic hepatitis B infection: the good and the bad. Lancet Gastroenterol. Hepatol. 5 , 406–417 (2020).
Maini, M. K. & Burton, A. R. Restoring, releasing or replacing adaptive immunity in chronic hepatitis B. Nat. Rev. Gastroenterol. Hepatol. 16 , 662–675 (2019).
Tighe, P. J., Ryder, R. R., Todd, I. & Fairclough, L. C. ELISA in the multiplex era: potentials and pitfalls. Proteom. Clin. Appl. 9 , 406–422 (2015).
Grifoni, A. et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181 , 1489–1501.e15 (2020).
Le Bert, N. et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. https://doi.org/10.1084/jem.20202617 (2021).
Weiskopf, D. et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. https://doi.org/10.1126/sciimmunol.abd2071 (2020).
Mazurek, G. H. & Villarino, M. E.; CDC. Guidelines for using the QuantiFERON-TB test for diagnosing latent Mycobacterium tuberculosis infection. Centers for Disease Control and Prevention. MMWR Recomm. Rep. 52 , 15–18 (2003).
Cornberg, M., Lok, A. S., Terrault, N. A. & Zoulim, F.; 2019 EASL-AASLD HBV Ttretment Endpoints Conference Faculty. Guidance for design and endpoints of clinical trials in chronic hepatitis B - Report from the 2019 EASL-AASLD HBV Treatment Endpoints Conference(double dagger). J. Hepatol. 72 , 539–557 (2020).
Gill, U. S. et al. Fine needle aspirates comprehensively sample intrahepatic immunity. Gut 68 , 1493–1503 (2019).
Gill, U. S., Pallett, L. J., Kennedy, P. T. F. & Maini, M. K. Liver sampling: a vital window into HBV pathogenesis on the path to functional cure. Gut 67 , 767–775 (2018).
Hartmann, F. J. & Bendall, S. C. Immune monitoring using mass cytometry and related high-dimensional imaging approaches. Nat. Rev. Rheumatol. 16 , 87–99 (2020).
Traum, D. et al. Highly multiplexed 2-dimensional imaging mass cytometry analysis of HBV-infected liver. JCI Insight https://doi.org/10.1172/jci.insight.146883 (2021).
Rendeiro, A. F. et al. The spatial landscape of lung pathology during COVID-19 progression. Nature 593 , 564–569 (2021).
Raimondo, G. et al. Statements from the Taormina expert meeting on occult hepatitis B virus infection. J. Hepatol. 49 , 652–657 (2008).
Caviglia, G. P. et al. Quantitation of HBV cccDNA in anti-HBc-positive liver donors by droplet digital PCR: a new tool to detect occult infection. J. Hepatol. 69 , 301–307 (2018).
Deguchi, M. et al. Evaluation of the highly sensitive chemiluminescent enzyme immunoassay “Lumipulse HBsAg-HQ” for hepatitis B virus screening. J. Clin. Lab. Anal. 32 , e22334 (2018).
Ozeki, I. et al. Analysis of hepatitis B surface antigen (HBsAg) using high-sensitivity HBsAg assays in hepatitis B virus carriers in whom HBsAg seroclearance was confirmed by conventional assays. Hepatol. Res. 48 , E263–E274 (2018).
Pepe, M. S. et al. Phases of biomarker development for early detection of cancer. J. Natl Cancer Inst. 93 , 1054–1061 (2001).
Kuhns, M. C. et al. Improved detection of early acute, late acute, and occult Hepatitis B infections by an increased sensitivity HBsAg assay. J. Clin. Virol. 118 , 41–45 (2019).
Liu, Y., Cathcart, A. L., Delaney, W. E. T. & Kitrinos, K. M. Development of a digital droplet PCR assay to measure HBV DNA in patients receiving long-term TDF treatment. J. Virol. Methods 249 , 189–193 (2017).
Musolino, C. et al. Behaviour of occult HBV infection in HCV-infected patients under treatment with direct-acting antivirals. Antivir. Ther. 24 , 187–192 (2019).
Kazemi-Shirazi, L., Petermann, D. & Muller, C. Hepatitis B virus DNA in sera and liver tissue of HBsAg negative patients with chronic hepatitis C. J. Hepatol. 33 , 785–790 (2000).
Kannangai, R. et al. Liver enzyme flares and occult hepatitis B in persons with chronic hepatitis C infection. J. Clin. Virol. 39 , 101–105 (2007).
Chemin, I., Guillaud, O., Queyron, P. C. & Trepo, C. Close monitoring of serum HBV DNA levels and liver enzymes levels is most useful in the management of patients with occult HBV infection. J. Hepatol. 51 , 824–825 (2009).
Saitta, C. et al. Risk of occult hepatitis B virus infection reactivation in patients with solid tumours undergoing chemotherapy. Dig. Liver Dis. 45 , 683–686 (2013).
Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7 , 6 (2021).
Yang, H. C. et al. Quantification of HBV core antibodies may help predict HBV reactivation in patients with lymphoma and resolved HBV infection. J. Hepatol. 69 , 286–292 (2018).
Kusumoto, S. et al. Ultra-high sensitivity HBsAg assay can diagnose HBV reactivation following rituximab-based therapy in patients with lymphoma. J. Hepatol. 73 , 285–293 (2020).
Parikh, N. D. et al. Biomarkers for the early detection of hepatocellular carcinoma. Cancer Epidemiol. Biomark. Prev. 29 , 2495–2503 (2020).
Chaiteerakij, R., Addissie, B. D. & Roberts, L. R. Update on biomarkers of hepatocellular carcinoma. Clin. Gastroenterol. Hepatol. 13 , 237–245 (2015).
Guidotti, L. G. et al. Viral clearance without destruction of infected cells during acute HBV infection. Science 284 , 825–829 (1999).
Xu, D., Su, C., Sun, L., Gao, Y. & Li, Y. Performance of serum Glypican 3 in diagnosis of hepatocellular carcinoma: a meta-analysis. Ann. Hepatol. 18 , 58–67 (2019).
Ge, T. et al. Diagnostic values of alpha-fetoprotein, dickkopf-1, and osteopontin for hepatocellular carcinoma. Med. Oncol. 32 , 59 (2015).
Johnson, P. J. et al. The detection of hepatocellular carcinoma using a prospectively developed and validated model based on serological biomarkers. Cancer Epidemiol. Biomark. Prev. 23 , 144–153 (2014).
von Felden, J., Garcia-Lezana, T., Schulze, K., Losic, B. & Villanueva, A. Liquid biopsy in the clinical management of hepatocellular carcinoma. Gut 69 , 2025–2034 (2020).
Wang, T. & Zhang, K. H. New blood biomarkers for the diagnosis of AFP-negative hepatocellular carcinoma. Front. Oncol. 10 , 1316 (2020).
Wong, G. L. et al. On-treatment alpha-fetoprotein is a specific tumor marker for hepatocellular carcinoma in patients with chronic hepatitis B receiving entecavir. Hepatology 59 , 986–995 (2014).
Marrero, J. A. Screening tests for hepatocellular carcinoma. Clin. Liver Dis. 9 , 235–251 (2005).
Tzartzeva, K. et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology 154 , 1706–1718.e1 (2018).
Gopal, P. et al. Factors that affect accuracy of alpha-fetoprotein test in detection of hepatocellular carcinoma in patients with cirrhosis. Clin. Gastroenterol. Hepatol. 12 , 870–877 (2014).
Simmons, O. et al. Predictors of adequate ultrasound quality for hepatocellular carcinoma surveillance in patients with cirrhosis. Aliment. Pharmacol. Therapeut. 45 , 169–177 (2017).
Del Poggio, P. et al. Factors that affect efficacy of ultrasound surveillance for early stage hepatocellular carcinoma in patients with cirrhosis. Clin. Gastroenterol. Hepatol. 12 , 1927–1933.e2 (2014).
Singal, A. G., Lampertico, P. & Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J. Hepatol. 72 , 250–261 (2020).
Loglio, A. et al. The combination of PIVKA-II and AFP improves the detection accuracy for HCC in HBV caucasian cirrhotics on long-term oral therapy. Liver Int. 40 , 1987–1996 (2020).
Park, S. J. et al. Usefulness of AFP, AFP-L3, and PIVKA-II, and their combinations in diagnosing hepatocellular carcinoma. Medicine 96 , e5811 (2017).
Lee, E., Edward, S., Singal, A. G., Lavieri, M. S. & Volk, M. Improving screening for hepatocellular carcinoma by incorporating data on levels of alpha-fetoprotein, over time. Clin. Gastroenterol. Hepatol. 11 , 437–440 (2013).
Choi, J. et al. Longitudinal assessment of three serum biomarkers to detect very early-stage hepatocellular carcinoma. Hepatology 69 , 1983–1994 (2019).
Kramvis, A. Genotypes and genetic variability of hepatitis B virus. Intervirology 57 , 141–150 (2014).
Bayliss, J. et al. Deep sequencing shows that HBV basal core promoter and precore variants reduce the likelihood of HBsAg loss following tenofovir disoproxil fumarate therapy in HBeAg-positive chronic hepatitis B. Gut 66 , 2013–2023 (2017).
Wong, D. et al. ALT flares during nucleotide analogue therapy are associated with HBsAg loss in genotype A HBeAg-positive chronic hepatitis B. Liver Int. 38 , 1760–1769 (2018).
Cornberg, M. & Glebe, D. Editorial: which factors influence HBsAg levels in HBV-infected patients? Aliment. Pharmacol. Therapeut. 52 , 547–548 (2020).
Rokuhara, A. et al. Hepatitis B virus RNA is measurable in serum and can be a new marker for monitoring lamivudine therapy. J. Gastroenterol. 41 , 785–790 (2006).
Wang, J. et al. Relationship between serum HBV-RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. J. Hepatol. https://doi.org/10.1016/j.jhep.2017.08.021 (2017).
Wang, J. et al. Reply to: “Serum HBV pgRNA as a clinical marker for cccDNA activity”: consistent loss of serum HBV RNA might predict the “para-functional cure” of chronic hepatitis B. J. Hepatol. 66 , 462–463 (2017).
Limothai, U. et al. Reverse transcriptase droplet digital PCR vs reverse transcriptase quantitative real-time PCR for serum HBV RNA quantification. J. Med. Virol. https://doi.org/10.1002/jmv.25792 (2020).
van Bommel, F. et al. Serum HBV RNA as a predictor of peginterferon Alfa-2a (40KD) response in patients with HBeAg-positive chronic hepatitis B. J. Infect. Dis. 218 , 1066–1074 (2018).
Tsuge, M. et al. Serum HBV RNA and HBeAg are useful markers for the safe discontinuation of nucleotide analogue treatments in chronic hepatitis B patients. J. Gastroenterol. 48 , 1188–1204 (2013).
Huang, Y. W. et al. On-treatment low serum HBV RNA level predicts initial virological response in chronic hepatitis B patients receiving nucleoside analogue therapy. Antivir. Ther. 20 , 369–375 (2015).
Laras, A., Koskinas, J., Dimou, E., Kostamena, A. & Hadziyannis, S. J. Intrahepatic levels and replicative activity of covalently closed circular hepatitis B virus DNA in chronically infected patients. Hepatology 44 , 694–702 (2006).
Scholtès, C. et al. Performance of a novel automated assay for the detection and quantification of HBV pregeomic RNA/ circulating RNAs in chronic HBV patients. Hepatology 72 , 447A (2020).
Loggi, E. et al. Serum hepatitis B core-related antigen is an effective tool to categorize patients with HBeAg-negative chronic hepatitis B. J. Viral Hepat. 26 , 568–575 (2019).
Lee, H. W. & Ahn, S. H. Prediction models of hepatocellular carcinoma development in chronic hepatitis B patients. World J. Gastroenterol. 22 , 8314–8321 (2016).
Weusten, J., Vermeulen, M., van Drimmelen, H. & Lelie, N. Refinement of a viral transmission risk model for blood donations in seroconversion window phase screened by nucleic acid testing in different pool sizes and repeat test algorithms. Transfusion 51 , 203–215 (2011).
European Association for the Study of the Liver. EASL 2017 Clinical Practice Guidelines on the management of hepatitis B virus infection. J. Hepatol. 67 , 370–398 (2017).
Vermeulen, M. et al. Hepatitis B virus transmission by blood transfusion during 4 years of individual-donation nucleic acid testing in South Africa: estimated and observed window period risk. Transfusion 52 , 880–892 (2012).
Ning, X. et al. Secretion of genome-free hepatitis B virus–single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog. 7 , e1002255 (2011).
Garcia, P. D., Ou, J. H., Rutter, W. J. & Walter, P. Targeting of the hepatitis B virus precore protein to the endoplasmic reticulum membrane: after signal peptide cleavage translocation can be aborted and the product released into the cytoplasm. J. Cell Biol. 106 , 1093–1104 (1988).
Ito, K., Kim, K. H., Lok, A. S. & Tong, S. Characterization of genotype-specific carboxyl-terminal cleavage sites of hepatitis B virus e antigen precursor and identification of furin as the candidate enzyme. J. Virol. 83 , 3507–3517 (2009).
Wang, J., Lee, A. S. & Ou, J. H. Proteolytic conversion of hepatitis B virus e antigen precursor to end product occurs in a postendoplasmic reticulum compartment. J. Virol. 65 , 5080–5083 (1991).
Thimme, R. et al. CD8 + T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J. Virol. 77 , 68–76 (2003).
Rehermann, B. Immune responses in hepatitis B virus infection. Semin. Liver Dis. 23 , 21–38 (2003).
Seto, W. K. et al. Hepatitis B reactivation in patients with previous hepatitis B virus exposure undergoing rituximab-containing chemotherapy for lymphoma: a prospective study. J. Clin. Oncol. 32 , 3736–3743 (2014).
Hakim, M. S., Spaan, M., Janssen, H. L. & Boonstra, A. Inhibitory receptor molecules in chronic hepatitis B and C infections: novel targets for immunotherapy? Rev. Med. Virol. 24 , 125–138 (2014).
Lopes, A. R. et al. Bim-mediated deletion of antigen-specific CD8 T cells in patients unable to control HBV infection. J. Clin. Invest. 118 , 1835–1845 (2008).
Peppa, D. et al. Up-regulation of a death receptor renders antiviral T cells susceptible to NK cell-mediated deletion. J. Exp. Med. 210 , 99–114 (2013).
Xu, D. et al. Circulating and liver resident CD4 + CD25 + regulatory T cells actively influence the antiviral immune response and disease progression in patients with hepatitis B. J. Immunol. 177 , 739–747 (2006).
Stoop, J. N. et al. Regulatory T cells contribute to the impaired immune response in patients with chronic hepatitis B virus infection. Hepatology 41 , 771–778 (2005).
Fisicaro, P. et al. Pathogenetic mechanisms of T cell dysfunction in chronic HBV infection and related therapeutic approaches. Front. Immunol. 11 , 849 (2020).
Pallett, L. J. et al. Metabolic regulation of hepatitis B immunopathology by myeloid-derived suppressor cells. Nat. Med. 21 , 591–600 (2015).
Fisicaro, P. et al. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B. Nat. Med. 23 , 327–336 (2017).
Salimzadeh, L. et al. PD-1 blockade partially recovers dysfunctional virus-specific B cells in chronic hepatitis B infection. J. Clin. Invest. 128 , 4573–4587 (2018).
Milich, D. & Liang, T. J. Exploring the biological basis of hepatitis B e antigen in hepatitis B virus infection. Hepatology 38 , 1075–1086 (2003).
Dunn, C. et al. Cytokines induced during chronic hepatitis B virus infection promote a pathway for NK cell-mediated liver damage. J. Exp. Med. 204 , 667–680 (2007).
Das, A. et al. Functional skewing of the global CD8 T cell population in chronic hepatitis B virus infection. J. Exp. Med. 205 , 2111–2124 (2008).
Sandalova, E. et al. Increased levels of arginase in patients with acute hepatitis B suppress antiviral T cells. Gastroenterology 143 , 78–87.e3 (2012).
Aiolfi, R. & Sitia, G. Chronic hepatitis B: role of anti-platelet therapy in inflammation control. Cell Mol. Immunol. 12 , 264–268 (2015).
Tiegs, G. & Lohse, A. W. Immune tolerance: what is unique about the liver. J. Autoimmun. 34 , 1–6 (2010).
Wohlleber, D. & Knolle, P. A. The role of liver sinusoidal cells in local hepatic immune surveillance. Clin. Transl. Immunol. 5 , e117 (2016).
Chang, K. M. et al. Distinct phenotype and function of circulating Vdelta1+and Vdelta2+gammadeltaT-cells in acute and chronic hepatitis B. PLoS Pathog. 15 , e1007715 (2019).
Boeijen, L. L. et al. Mucosal-associated invariant T cells are more activated in chronic hepatitis B, but not depleted in blood: reversal by antiviral therapy. J. Infect. Dis. 216 , 969–976 (2017).
Yoshio, S. et al. Indoleamine-2,3-dioxygenase as an effector and an indicator of protective immune responses in patients with acute hepatitis B. Hepatology 63 , 83–94 (2016).
Hou, F. Q. et al. Rapid downregulation of programmed death-1 and interferon-gamma-inducible protein-10 expression is associated with favourable outcome during antiviral treatment of chronic hepatitis B. J. Viral Hepat. 20 (Suppl. 1), 18–26 (2013).
Tan, A. T. et al. A longitudinal analysis of innate and adaptive immune profile during hepatic flares in chronic hepatitis B. J. Hepatol. 52 , 330–339 (2010).
Wang, Y. et al. Predictive value of interferon-gamma inducible protein 10 kD for hepatitis B e antigen clearance and hepatitis B surface antigen decline during pegylated interferon alpha therapy in chronic hepatitis B patients. Antivir. Res. 103 , 51–59 (2014).
Chen, Y. et al. Development of a sandwich ELISA for evaluating soluble PD-L1 (CD274) in human sera of different ages as well as supernatants of PD-L1 + cell lines. Cytokine 56 , 231–238 (2011).
Cheng, H. Y. et al. Circulating programmed death-1 as a marker for sustained high hepatitis B viral load and risk of hepatocellular carcinoma. PLoS One 9 , e95870 (2014).
Zhou, L. et al. Soluble programmed death-1 is a useful indicator for inflammatory and fibrosis severity in chronic hepatitis B. J. Viral Hepat. 26 , 795–802 (2019).
Jeng, W.-J. & Yang, H.-I. Discrepant range of sPD-1 in different studies of chronic hepatitis B. A letter in response to soluble programmed death-1 is a useful indicator for inflammatory and fibrosis severity in chronic hepatitis B. J. Viral Hepat. 26 , 930–931 (2019).
Jaroszewicz, J. et al. Hepatitis B surface antigen (HBsAg) decrease and serum interferon-inducible protein-10 levels as predictive markers for HBsAg loss during treatment with nucleoside/nucleotide analogues. Antivir. Ther. 16 , 915–924 (2011).
Sonneveld, M. J., Arends, P., Boonstra, A., Hansen, B. E. & Janssen, H. L. Serum levels of interferon-gamma-inducible protein 10 and response to peginterferon therapy in HBeAg-positive chronic hepatitis B. J. Hepatol. 58 , 898–903 (2013).
Yoshio, S. et al. Cytokine and chemokine signatures associated with hepatitis B surface antigen loss in hepatitis B patients. JCI Insight https://doi.org/10.1172/jci.insight.122268 (2018).
Johnson Valiente, A. et al. The inflammatory cytokine profile associated with liver damage is broader and stronger in patients with chronic hepatitis B compared to patients with acute hepatitis B. J. Infect. Dis. 225 , 470–475 (2022).
Xia, J. et al. Profiles of serum soluble programmed death-1 and programmed death-ligand 1 levels in chronic hepatitis B virus-infected patients with different disease phases and after anti-viral treatment. Aliment. Pharmacol. Therapeut. 51 , 1180–1187 (2020).
Dou, Y. et al. Elevated serum levels of soluble CD14 in HBeAg-positive chronic HBV patients upon Peginterferon treatment are associated with treatment response. J. Viral Hepat. 26 , 1076–1085 (2019).
Sandler, N. G. et al. Host response to translocated microbial products predicts outcomes of patients with HBV or HCV infection. Gastroenterology 141 , 1220–1230 (2011).
Download references
Acknowledgements
The authors thank the Workshop Participants: The ICE-HBV HBV Serum Biomarkers Workshop was held virtually in two sessions on 5th and 12th October 2020 ( https://ice-hbv.org/hbv-serum-biomarkers-workshop/ ). The chairs of the workshop A.K. and P.R. organized the meeting with C.P. K.M.C., M.D., P.F., D.G., J.H., H.L.A.J., D.T.Y.L., T.P., B.T. and F.V.P. presented at the workshop. O.A., M.B.M., T.M.B., H.L.Y.C., G.A.C., W.D., A.M.G., A.G., O.L., M.M., V.M., U.P., J.Y., M.F.Y. and F.B. chaired the workshop sessions and/or participated in the panel discussions. The authors thank T. Candy (VIDRL, RMH, Doherty Institute, Melbourne, Victoria, Australia) for assistance with the preparation of the manuscript.
Author information
Authors and affiliations.
Hepatitis Virus Diversity Research Unit, Department of Internal Medicine, School of Clinical Medicine, University of the Witwatersrand, Johannesburg, South Africa
Anna Kramvis
The Corporal Michael J. Crescenz Veterans Affairs Medical Center and University of Pennsylvania Perelman School of Medicine, Philadelphia, PA, USA
Kyong-Mi Chang
Department of Internal Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany
Maura Dandri
German Centre for Infection Research (DZIF), Hamburg-Lübeck-Borstel-Riems partner site, Hamburg, Germany
Hepatic Pathogenesis Section, Laboratory of Infectious Diseases, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD, USA
Patrizia Farci
National Reference Center for Hepatitis B Viruses and Hepatitis D Viruses, Institute of Medical Virology, Justus Liebig University Giessen, Giessen, Germany
Dieter Glebe
German Center for Infection Research (DZIF), Partner Site Giessen-Marburg-Langen, Giessen, Germany
Department of Microbiology and Immunology, The Pennsylvania State University College of Medicine, Philadelphia, PA, USA
Jianming Hu
Toronto Centre for Liver Disease, University of Toronto, Toronto, Canada
Harry L. A. Janssen
Division of Gastroenterology and Hepatology, Department of Medicine, Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts, USA
Daryl T. Y. Lau
Peter Doherty Institute for Infection and Immunity, Melbourne, Australia
Capucine Penicaud
Laboratory of Molecular Hepatology, Department of Human Pathology, University Hospital “G. Martino” of Messina, Messina, Italy
Teresa Pollicino
INSERM U1052, CNRS UMR-5286, Cancer Research Center of Lyon (CRCL), Lyon, France
Barbara Testoni
University of Lyon, Université Claude-Bernard (UCBL), Lyon, France
Department of Hepatology, Leipzig University Medical Center, Leipzig, Germany
Florian Van Bömmel
Basic Medical Sciences, Purdue University, West Lafayette, Indiana, USA
Ourania Andrisani
Janssen Infectious Diseases, Janssen, Titusville, USA
Maria Beumont-Mauviel
Baruch S. Blumberg Institute, Doylestown, Pennsylvania, USA
Timothy M. Block
Chinese University of Hong Kong, Shatin, Hong Kong
Henry L. Y. Chan
Union Hospital, Shatin, Hong Kong
ID Core Research, Abbott Laboratories, Abbott Park, IL, USA
Gavin A. Cloherty
Assembly Bio, South San Francisco, San Francisco, CA, USA
William E. Delaney
Roche Pharma Research & Early Development, Basel, Switzerland
Anna Maria Geretti
Department of Infectious Diseases, Fondazione PTV, Faculty of Medicine, University of Rome Tor Vergata, Rome, Italy
Department of Infectious Diseases, School of Immunology & Microbial Sciences, King’s College London, London, UK
Toronto Centre for Liver Disease, University Health Network, Toronto, Canada
Adam Gehring
Victorian Infectious Diseases Reference Laboratory, Royal Melbourne Hospital at the Peter Doherty Institute for Infection and Immunity, Melbourne, Australia
Kathy Jackson & Peter A. Revill
Janssen Pharmaceutica, Beerse, Belgium
Oliver Lenz
Division of Infection & Immunity, Institute of Immunity & Transplantation, University College London, London, UK
Mala K. Maini
Forum for Collaborative Research, University of California Berkeley School of Public Health, Washington DC Campus, Washington, DC, USA
Veronica Miller
Institute of Virology, School of Medicine, Technical University of Munich, Helmholtz Zentrum München, Munich, Germany
Ulrike Protzer
Gilead Sciences, Foster City, CA, USA
Jenny C. Yang
Department of Medicine, Queen Mary Hospital, The University of Hong Kong, Hong Kong, China
Man-Fung Yuen
State Key Laboratory of Liver Research, The University of Hong Kong, Hong Kong, China
INSERM Unit 1052 – Cancer Research Center of Lyon, Hospices Civils de Lyon, Lyon University, Lyon, France
Fabien Zoulim
Department of Microbiology and Immunology, University of Melbourne, Parkville, Victoria, Australia
Peter A. Revill
You can also search for this author in PubMed Google Scholar
Contributions
P.R., A.K., K.M.C., M.D., P.F., D.G., J.H., H.L.J., D.L., M.C.P., T.P., B.T., F.B., O.M.A., M.B.M., T.B., H.L.Y.C., G.A.C., W.E.D., A.M.A.G., A.J.G., K.J., O.L., M.K.M., V.M., U.P., J.C.Y., M.F.Y. and F.Z. researched data for the article. P.R., A.K., K.M.C., M.D., P.F., D.G., J.H., H.L.J., D.L., M.C.P., T.P., B.T., F.B., O.M.A., M.B.M., T.B., H.L.Y.C., G.A.C., W.E.D., A.M.A.G., A.J.G., K.J., O.L., M.K.M., V.M., U.P., J.C.Y., M.F.Y. and F.Z. contributed substantially to discussion of the content. P.R., A.K., K.M.C., M.D., P.F., D.G., J.H., H.L.J., D.L., M.C.P., T.P., B.T., F.B., O.M.A., M.B.M., T.B., H.L.Y.C., G.A.C., W.E.D., A.M.A.G., A.J.G., O.L., M.K.M., V.M., U.P., J.C.Y., M.F.Y. and F.Z. wrote the article. P.R., A.K., K.M.C., M.D., P.F., D.G., J.H., H.L.J., D.L., M.C.P., T.P., B.T., F.B., O.M.A., M.B.M., T.B., H.L.Y.C., G.A.C., W.E.D., A.M.A.G., A.J.G., K.J., O.L., M.K.M., V.M., U.P., J.C.Y., M.F.Y. and F.Z. reviewed and/or edited the manuscript before submission.
Corresponding authors
Correspondence to Anna Kramvis or Peter A. Revill .
Ethics declarations
Competing interests.
A.K. is a recipient of a grant from the Cancer Association of South Africa (CANSA). K.M.C. is supported by the Corporal Michael J. Crescenz VA Medical Center Research Program in Philadelphia, Pennsylvania 19104, USA and has served in the Scientific Advisory Committee for Arbutus Biopharma. M.D. is supported by the German Research Foundation (DFG; SFB841), the German Center for Infection Research (DZIF) and the Dandri lab has received collaborative funding from Gilead Sciences and MYR-GmbH. P.F. has nothing to declare and is supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MA, USA. D.G. is supported by the German Research Foundation (DFG; SFB1021), the German Center for Infection Research (DZIF), the Robert Koch Institute, Berlin and the German Federal Ministry of Health. J.H. has been supported by funding from the National Institute of Allergy and Infectious Disease/NIH and Gilead for work relevant here and has consulted for Arbutus, Bristol-Myers-Squibb, Gilead, Janssen, Roche and Sanofi. H.L.A.J. received grants from AbbVie, Gilead Sciences, GlaxoSmithKline, Janssen, Roche, Vir Biotechnology Inc. and is a consultant for Aligos, Antios, Arbutus, Eiger, Gilead Sciences, GlaxoSmithKline, Janssen, Merck, Roche, VBI Vaccines, Vir Biotechnology Inc. and Viroclinics. D.T.Y.L. received research grants from GlaxoSmithKline, Janssen and Abbott Laboratories. C.P. is a consultant for Janssen. T.P. has been a speaker for Gilead Science. B.T. has nothing to declare. F.V.B. has received research grants from Gilead Sciences, Roche Diagnostics, Ipsen and Janssen; has been part of speaker’s bureau for Gilead Sciences, Roche, Janssen, Ipsen, Eisai, MSD and GSK; and has received support for conference travels from Gilead, Janssen, Roche, Ibsen, MSD and Bayer. O.A. has nothing to disclose, supported by 5R01DK044533-23. M.B.M. is an employee of Janssen Pharmaceuticals. T.M.B. is on the Board of Hepion Pharma and has received support from Arbutus Biopharma and is a co-founder and equity holder in Glycotest. H.L.Y.C. has served as an adviser of AbbVie, Aligos, Arbutus, Gilead, GSK, Hepion, Janssen, Merck, Roche, Vaccitech, VenatoRx, Vir Biotechnology and Virion Therapeutics, and is a speaker for Gilead, Mylan and Roche. G.C. is an Abbott Employee and shareholder. W.E.D. is an employee of and owns stock in Assembly Bio and owns stock in Gilead Sciences. A.M.G. is an employee of Roche Pharma Research and Early Development and also holds stock units with the company. A.G. receives research funding from Janssen Pharmaceuticals, GSK, and Gilead Sciences and conducts consulting/scientific advising for Janssen Pharmaceuticals, Roche, GSK, Vir Biotech, Finch Therapeutics and SQZ Biotech. O.L. is an employee of Janssen Pharmaceutical NV and owns stock of Johnson and Johnson. K.J. performs contract research for Gilead Sciences and Arrowhead Pharmaceuticals. The M.K.M. lab has received collaborative funding from Gilead Sciences, VIR Biotechnology, Hoffmann-La-Roche and GSK (last 3 years), with no funds taken personally. M.K.M. is supported by Wellcome Investigator Award 21419/Z/18/Z. V.M. and the Forum for Collaborative Research, University of California Berkeley School of Public Health, Washington DC Campus, Washington, DC, USA: the Forum received unrestricted support from multiple companies, but did not receive funding specific to the writing of this manuscript. The companies are: Abbott Diagnostics, Aligos Therapeutics, Inc., Altimmune, Antios, Therapeutics, Assembly Biosciences, Eiger Biopharmaceuticals, ENYO Pharma, Gilead, GSK, Immunocore, Janssen Pharmaceuticals ID&V, Monogram Biosciences Quest Diagnostics, Roche Pharma R&D (pRED), Venatorx Pharmaceuticals, Inc., Vir Biotechnology, Virion Therapeutics, LLC, Viroclinics-DDL Diagnostic Laboratory. U.P. is co-founder and shareholder of SCG Cell Therapy, obtained research support from Abbott, ALIOS, Yhlo and VirBio, and received personal fees from Abbvie, Arbutus, Gilead, GSK, J&J, Roche, Sanofi, Sobi and Vaccitech. J.C.Y. was an employee of Gilead Sciences. M.F.Y. reports being an adviser/consultant for and/or having received grant/research support from AbbVie, Aligos Therapeutics, Antios Therapeutics, Arbutus Biopharma, Arrowhead Pharmaceuticals, Assembly Biosciences, Bristol-Myers Squibb, Dicerna Pharmaceuticals, Finch Therapeutics, Fujirebio Incorporation, GlaxoSmithKline, Gilead Sciences, Immunocore, Janssen, Merck Sharp and Dohme, Clear B Therapeutics, Springbank Pharmaceuticals, Silverback Therapeutics, Sysmex Corporation, Vir Biotechnology and Roche. F.Z. reports consulting for Aligos, Antios, Arbutus, Assembly, Enochian, Gilead, GSK, Roche Molecular Systems, and Zhimeng and research funding to INSERM from Assembly, Beam and Viravaxx. P.A.R. has previously received research funding from Gilead Sciences and is on the Scientific Advisory Board of Enochian Biosciences.
Peer review
Peer review information.
Nature Reviews Gastroenterology & Hepatology thanks Jia-Horng Kao, Philippa Matthews and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
HBV Forum: https://forumresearch.org/hbv-forum/hbv-foruman-overview
Hepatitis B Foundation: https://www.hepb.org/
ICE-HBV: https://ice-hbv.org/
Workshop on HBV biomarkers: https://ice-hbv.org/hbv-serum-biomarkers-workshop/
Supplementary information
Supplementary information, rights and permissions.
Reprints and permissions
About this article
Cite this article.
Kramvis, A., Chang, KM., Dandri, M. et al. A roadmap for serum biomarkers for hepatitis B virus: current status and future outlook. Nat Rev Gastroenterol Hepatol 19 , 727–745 (2022). https://doi.org/10.1038/s41575-022-00649-z
Download citation
Accepted : 16 June 2022
Published : 20 July 2022
Issue Date : November 2022
DOI : https://doi.org/10.1038/s41575-022-00649-z
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Microrna levels in patients with chronic hepatitis b virus and hiv coinfection in a high-prevalence setting; kwazulu-natal, south africa.
- Lulama Mthethwa
- Raveen Parboosing
- Nokukhanya Msomi
BMC Infectious Diseases (2024)
Prevalence and influential factors of isolated hepatitis B core antibody positivity in a Chinese adult population
- Chengwei Wang
- Jianchun Xian
Scientific Reports (2024)
The hijacking of HBV by small extracellular vesicles inhibits M1 macrophages to facilitate immune evasion
- Wanlong Pan
A two-pronged strategy utilizing exosomes extracted from antigen-presenting cells to combat hepatitis B
- Dangyang Wang
Nano Research (2024)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- v.13(10); 2021 Oct

Critical Updates on Chronic Hepatitis B Virus Infection in 2021
Cyriac a philips.
1 Clinical and Translational Hepatology, The Liver Institute, Rajagiri Hospital, Aluva, IND
Rizwan Ahamed
2 Gastroenterology and Advanced Gastrointestinal Endoscopy, Center of Excellence in Gastrointestinal Sciences, Rajagiri Hospital, Aluva, IND
Jinsha K Abduljaleel
Sasidharan rajesh.
3 Diagnostic and Interventional Radiology, Center of Excellence in Gastrointestinal Sciences, Rajagiri Hospital, Aluva, IND
Philip Augustine
Chronic hepatitis B virus (HBV) infection is a global healthcare burden in the form of chronic liver disease, cirrhosis, liver failure and liver cancer. There is no definite cure for the virus and even though extensive vaccination programs have reduced the burden of liver disease in the future population, treatment options to eradicate the virus from the host are still lacking. In this review, we discuss in detail current updates on the structure and applied biology of the virus in the host, examine updates to current treatment and explore novel and state-of-the-art therapeutics in the pipeline for management of chronic HBV. Furthermore, we also specifically review clinical updates on HBV-related acute on chronic liver failure (ACLF). Current treatments for chronic HBV infection have seen important updates in the form of considerations for treating patients in the immune tolerant phase and some clarity on end points for treatment and decisions on finite therapy with nucleos(t)ide inhibitors. Ongoing cutting-edge research on HBV biology has helped us identify novel target areas in the life cycle of the virus for application of new therapeutics. Due to improvements in the area of genomics, the hope for therapeutic vaccines, vector-based treatments and focused management aimed at targeting host integration of the virus and thereby a total cure could become a reality in the near future. Newer clinical prognostic tools have improved our understanding of timing of specific treatment options for the catastrophic syndrome of ACLF secondary to reactivation of HBV. In this review, we discuss in detail pertinent updates regarding virus biology and novel therapeutic targets with special focus on the appraisal of prognostic scores and treatment options in HBV-related ACLF.
Introduction and background
The prevalence of hepatitis B virus (HBV) surface antigen positivity among the general population differs according to geographical region, which also dictates possible routes of transmission. In low prevalence (<2%) regions such as North America and Western Europe, age at infection in early adulthood and route of transmission is mostly sexual and percutaneous. In regions of moderate prevalence (2-8%), age at infection in childhood and perinatal transmission is the most common mode of spread. Previously, age-dependent phases of HBV were described as immunotolerant phase (high replication, low-inflammation), immunoactive phase, inactive carrier state (low replication levels with normal/nearly normal serum aminotransferase levels) and reactivation. Nonetheless, these have been renamed recently as HBV envelope antigen (HBeAg)-positive infection, HBeAg-positive hepatitis and HBeAg-negative infection and HBeAg-negative hepatitis. Progression to cirrhosis in HBeAg-positive patients occurs at a rate of 2 to 5.5% per year increasing from 8 to 20% in five years. Inactive carriers who have normal aminotransferase levels and HBV DNA levels <2,000 IU/ml experience disease regression at the rate of 0.5 to 2% per year. HBeAg-negative hepatitis, or the reactivation phase, represents a progressive stage of chronic HBV. Anti-HBe (antibody to e-antigen)-positive patients experience rapid progression to cirrhosis at an annual rate of 8 to 20%. Patients with cirrhosis progress to advanced liver disease and hepatic failure at a rate of 16% over five years [ 1 - 4 ]. Chronic HBV infection remains a huge burden on the patients, their family and the healthcare system the world over, mostly in the Asia-Pacific region. There have been rapid developments toward a functional cure of HBV infection, with novel compounds currently in various study phases. Our current understanding of pathogenesis, immunology and clinical outcomes of HBV infection has seen vast updates over the last decade. In this narrative review, we provide in-depth discussions on the current understanding of biology and immuno-pathogenesis; variants and genotypes of HBV infection and extrapolate the same toward discussing novel therapies. We also explore current treatment options and discuss with clarity the guideline recommendations on HBV treatments, specifically updates on the special clinical condition of acute on chronic liver failure (ACLF) related to HBV infection.
Updates on HBV-related applied biology
The HBV Structure
HBV infection is a dynamic disease that encompasses biochemical, histological and clinical changes that occur over time, depending on the mode of acquisition, host and environmental factors. Within the host, HBV can exist in three forms, the infectious virion (Dane particle) and non-infectious particles that include enveloped nucleocapsids containing immature DNA/RNA, subviral particles (spheres, filaments lacking nucleocapsid proteins) and naked nucleocapsids [ 5 , 6 ]. According to the Baltimore Classification, a system utilized to group viruses taking into consideration both transcription and replication, on the basis of manner of messenger RNA (mRNA) synthesis, HBV belongs to Group VII which includes double-stranded DNA viruses with an RNA intermediate. HBV is a partially double-stranded hepadnavirus with a size of 42 nm containing a relaxed-circular DNA (rcDNA) genome with complete minus and incomplete plus strands. It has a host-derived outer surface lipid coat containing surface antigen which consists of large (L-), middle (M-) and small (S-HBsAg) and an inner core protein called the hepatitis B core antigen (HBcAg). The pre-S1 domain of the L-HBs plays a key role in viral envelopment and drives infectivity [ 7 ].
The viral genome encodes four overlapping open reading frames (ORFs): C (pre-core and core regions), P, S (pre-S1, pre-S2, S regions), and X (from which functional viral proteins are produced). The core antigen (nucleocapsid) protein, HBcAg; and the ‘e’ antigen (HBeAg) and 22-kDa pre-core protein (p22cr) are produced from ORF-C core and pre-core regions respectively. The polymerase protein (Pol) encoded from ORF-P is made of terminal domain with functions of encapsidation and initiation of minus strand synthesis; the reverse transcriptase domian (RT) which catalyzes genome synthesis; and the ribonuclease (H) domain which degrades pregenomic RNA and facilitates replication. HBV X antigen protein (HBxAg) is encoded by ORF-X and has multiple functions that support various stages of viral replication including signal transduction, DNA repair, activation of transcription pathways and inhibition of protein degradation along with participation in the oncogenic potential of HBV [ 8 - 11 ]. In the S-domain, the intermolecular disulfide bonds contribute to the structural stability of spherical virions and promote high resistance of HBV to inactivation by dehydration and heat stress [ 12 , 13 ]. Other important functional elements include direct repeats (DR1 and DR2) required for strand-specific synthesis of DNA during replication and enhancer elements (En1 and En2) which promote liver-specific expression of HBV gene products. Apart from this, a glucocorticoid-responsive element (GRE) sequence within the S-domain, a post-transcriptional regulatory element also exists. This region controls gene transcription and protein activation either by reversible events such as posttranslational modifications of phosphorylation or sequestration; and via irreversible events such as proteolysis. The GRE overlaps En1 and the polyadenylation signal (which makes transcribed RNA more stable, prevents degradation and allows the mature messenger RNA molecule to be exported from the nucleus and translated into a protein by ribosomes in the cytoplasm) within the core gene [ 14 ]. To summarize, apart from the major structural proteins, additional functional components in HBV have been demonstrated to enhance replication, promote liver-specific expression of viral proteins, prevent viral protein degradation and improve structural stability during cellular entry.
Updates on Viral Entry
The mode of entry and HBV replication steps within the hepatocyte has been extensively updated in the last decade. The virus attaches to the host cell surface (basolateral membrane of hepatocyte) through initial low-affinity binding on highly sulfated-heparan sulfate proteoglycans (HSPG) such as hepatotropic glypican-5 followed by high-affinity binding on target receptor. This binding to the HSPGs is mediated by electrostatic interactions between the negatively charged HSPG and two positively charged residues of the S-domain [ 15 ]. Initial studies showed that heparin, a glycosaminoglycan, interfered with HBV attachment. The higher the sulfation, the stronger the inhibition - lesser sulfated glycosaminoglycans such as chondroitin sulfate were less effective in blocking HBV entry. Thereafter the region between amino acids at position 2 and 47 of the pre-S1 of the HBV acts as receptor binder and attaches to the liver cell entry receptor. The latter was identified as the sodium taurocholate co-transporting peptide (NTCP, coded by the SLC10A1 gene; functions to uptake conjugated bile acids into hepatocytes). It is interesting to note that NTCP expression is rapidly lost after isolation of primary human hepatocytes and is absent in poorly differentiated hepatocellular carcinomas (HCC). Thus, malignant hepatoma cells and primary hepatocytes do not support and are not susceptible to infection with HBV (lack of efficient cell culture system permissive for viral infection and replication). Nonetheless, recently, human pluripotent stem cells transformed to hepatocyte-like cells (HLC) were found capable of expressing hepatocyte markers and host factors needed for the development of HBV infection [ 16 ]. Virus internalization into the hepatocyte cytoplasm occurs through the process of endocytosis in which the viral material to be internalized is surrounded by an area of host cell plasma membrane, which then buds off inside the cell to form a vesicle containing the ingested viral material. HBV infection was low in cell lines with overexpressed NTCP. This meant that the co-factors for internalization and infection were important for viral infection. It was identified that the receptor tyrosine kinase, also known as the epidermal growth factor receptor (EGFR), through interaction with the NTCP triggers the internalization and endocytosis process mediated by the caveolae-1/lipid-raft and possibly clathrin, leads to the formation of endosomes in the cytoplasm [ 17 , 18 ]. The host-cell protein, the calcium-dependent cell adhesion E cadherin was shown to play a central role in HBV entry. This protein binds to the glycosylated NTCP and promotes relocation to the basolateral membrane (cell polarization). On a different note, the cell-polarization limits entry of hepatitis C virus through tight junctions that restrict viral access to receptor binding [ 19 ].
There are three different types of endosomes: early endosomes, late endosomes, and recycling endosomes, differentiated by their morphology, the time taken for the endocytosed material to reach them, and by markers such as Ras superfamily of G proteins called Rabs. Once endocytic vesicles uncoat, they fuse with early endosomes (via Rab5A) which then mature into late endosomes before fusing with lysosomes (via Rab7A) [ 20 ]. In the HBV internalization cycle, the EGFR activation triggers a time-dependent relocalization of HBV pre-S1 to early and late endosomes and to lysosomes in concert with EGFR transport. However, blockade of EGFR-downstream signaling proteins including mitogen-activated protein kinase (MAPK), phosphoinositide 3-kinase (PI3K), and signal transducer and activator of transcription (STAT), does not have a significant effect in reducing HBV infection. Interestingly, efficiency of EGFR endocytosis and HBV entry were reduced when there was a deleterious mutation in EGFR or genetic knockdown of endocytosis adaptor molecules. In this regard, it was demonstrated that the suppression of EGFR ubiquitination by site-directed mutagenesis or knockdown of the EGFR-sorting molecules [signal-transducing adaptor molecule (STAM) and lysosomal protein transmembrane 4β (LAPTM4B)] ameliorated EGFR transport to the late endosome which was shown to be critical for efficient HBV infection. Another novel finding is that the hepatocyte NTCP undergoes extensive oligomerization in the presence of HBV preS1. Oligomerization refers to the interaction of more than one polypeptide chain, which results in the formation of the quaternary structure, generally considered to be the highest level of organization within the protein structural hierarchy. The drug troglitazone (but not pioglitazone) blocked internalization of HBV preS1 and its receptor, NTCP by preventing oligomerization. This work represented the importance of small molecule and peptide-based therapy in prevention of HBV infection [ 21 , 22 ]. Clathrin-mediated virus entry also plays a role in HBV internalization, in which the interaction with protein adapter-2 (AP-2) facilitates infection. Silibinin, a known inhibitor of clathrin-mediated endocytosis was shown to reduce HBV virus entry of HepG2-NTCP cell line [ 23 ]. Internalized virus escapes from the endocytic pathway once signaling that support fusion is activated. The crucial aspect in fusion mechanism is dependent on the pH. Some of the identified (but not confirmed) fusogenic domains include the C-terminal half of the pre-S2 region, the N-terminal of the S-region, pre-S1 region and the N-terminal of pre-S1 region [ 24 , 25 ]. To summarize, HBV entry into hepatocytes is not only governed by attachment of pre-S1 to the NTCP receptor, but also initial priming through low-affinity binding with heparan sulfate proteins on hepatocyte, internalization via the tyrosine receptor kinase EGFR, oligomerization to stabilize viral structure orchestrated by multiple other small molecules such as host cell calcium-dependent E cadherin, clathrin and adapter proteins that all form novel drug targets. For example, bafilomycin A1, an inhibitor of vacuolar enzymes responsible for acidification of pH gradient within endosomes inhibited HBV in duck hepatocytes and ameliorated HBV in human cell lines. Further, silencing of small molecules (Rabs) that transport plasma membranes to endosomes also significantly reduced HBV infection. Depending on the structure and biology of HBV, various entry inhibitors have been tested in pre-clinical studies. These include: a. Attachment inhibitors targeting S-,M-,L-HBs (heparin and suramin) or pre-S1 (proanthocyanidins); b. HSPGs (synthetic anti-lipopolysaccharide peptides); c. Substrate inhibitors of NTCP that target NTCP [taurocholic acid and derivatives such as ursodeoxycholic acid (UDCA), tauro-UDCA and glyco-UDCA, irbesartan]; d. Those targeting NTCP + Niemann-Pick C1-Like 1 (NPC1L1) protein (ezetimibe); e. Direct inhibitors of NTCP that interfere (cyclosporine A, vanitaracin A); f. Those that mildly or do not interfere (myrcludex-B, SCY450, SCY995 and Evans blue) with bile acid uptake and; g. Those which directly regulate NTCP expression (Ro41-5253, retinoic acid receptor antagonist) [ 26 , 27 ].
Updates on Nuclear Transport, Assembly and Release
After escape from the late endosome, the viral particles traverse the cytoplasm towards the host cell nucleus. As previously stated, the membrane fusion leads to direct release of nucleocapsids into the cytoplasm. A conserved membrane-permeable peptide within the surface protein of HBV was recently identified, of the pre-S2 domain, called the translocation motif (TLM). The TLM promotes delivery of proteins and nucleic acids into cells and tissues. Surface exposure of TLM peptides on the HBV surface protein due to proteolytic processing leads to fusion of peptides to HBc protein enabling formation of fully assembled capsids [ 28 - 32 ]. These viral capsids then translocate (via microtubule mediated transport) as complete ‘virus’ across cytoplasm towards the nucleus. The microtubule transport assembly is dependent on tubulin distribution and linkage of capsids to the dynein-motor-complex (cytoskeletal motor proteins that move along microtubules). Nocodazole is a drug that can depolymerize microtubules and thus block viral nucleocapsids from reaching the host cell nucleus, preventing formation of covalently closed circular DNA (cccDNA) that defines the HBV life cycle. Nonetheless, microtubule destabilizing drugs are associated with severe side effects and cannot be utilized in clinical setting [ 33 - 36 ]. The viral nucleocapsids undergo disassembly at the host-cell nuclear pore complex where the HBV rcDNA is converted to cccDNA which serves as a template for transcription of viral RNAs (pre-genomic and sub-genomic RNA). HBV pre-genomic RNA contains a stem loop called epsilon which is essential for RNA generation and packaging into viral capsids. It is through interaction with the Zinc finger antiviral protein (ZAP), interferon treatment destabilizes RNA generation and reduces viral replication. Similar to ZAP, recent studies have identified that multiple other small molecules and cellular factors interact with the HBV RNA to promote or suppress degradation and affect viral replication. These include cytidine deaminase, splicing factors, small ribonucleoprotein, RNA-binding motif protein and peroxiredoxins, which also act as small molecular targets for HVB therapy [ 37 - 40 ].
Updates on Viral Transcription and cccDNA
Within the cytoplasm, along with viral polymerase, pre-genomic RNA is encapsidated by HBV core protein to form a viral capsid. Inside the viral capsid, pre-genomic RNA undergoes reverse transcription to generate single-stranded negative-strand DNA (immature nucleocapsids, within infected cells), further followed by generation of partially double-stranded DNA (mature nucleocapsids, in released viral particles) yielding viral rcDNA. These capsids containing rcDNA are either transported back into the nucleus to increase the cccDNA pool or enveloped and released as progeny virions. HBsAg production is predominantly from cccDNA in younger HBeAg-positive patients. Hypo-phosphorylation of capsid proteins produces regular virions while hyper-phosphorylation produces empty virions [ 41 - 44 ]. The reverse transcription also produces aberrant by-products called HBV double-stranded linear DNA that are either released as defective virions or integrate with the host genome. This aberrant integration fails to transcribe pre-genomic RNA (no replicative power), but could still act as a template for generation of HBsAg. This happens in older chronic HBV patients who are HBeAg negative. Currently approved medications for HBV such as interferon-α and nucleos(t)ide reverse transcriptase inhibitors (NAs) reduce viral replication and slow disease progression, but do not cure chronic HBV infection. This is because these agents do not have any effect on persistent HBV cccDNA, since cccDNA formation is not only dependent on the viral DNA polymerase but also the host DNA polymerase(s). In this regard, it was shown that the anti-retroviral host factor SAMHD1 binds to single-stranded virus DNA, acting as a scaffolding protein to facilitate formation of cccDNA through relaxed circular DNA repair processes [ 45 - 47 ]. The HBx protein was demonstrated to activate HBV transcription through recruitment onto cccDNA. HBx also counteracts host restriction mechanisms of cccDNA transcription. Recently, the smallest known proteins with prolyl isomerase activity, which catalyze the cis-trans isomerization of proline peptide bonds, Parvulin 14 and Parvulin 17, were discovered to bind to HBx and cccDNA and promote HBV replication in an HBx-dependent manner. Thus, HBx itself and HBx-involved protein-protein interactions form novel molecular targets for therapeutic development against HBV [ 48 - 50 ]. A recent study found that the Smc5/6 of the structural maintenance of chromosomes family suppresses HBV replication. The drug nitazoxanide was found to block the inhibitor of Smc5/6 (damage specific DNA binding protein 1 or DDB1 binding to HBx protein) and promote suppression of replication [ 51 ]. Each infected hepatocyte contains one to 10 cccDNA copies with a half-life of 9.2 months in NA-treated chronic HBV patients. To clear cccDNA from infected cells, apart from direct targeting of cccDNA, two other steps are imperative. First, viral replication and cccDNA replenishment must be completely blocked, and, second, exhaustion of the pool of pre-existing cccDNA within a specified time frame. In the presence of potent suppression of viral replication with an NA addition of small interfering RNA or capsid inhibitor may help clear cccDNA completely [ 52 - 54 ].
With respect to cccDNA clearance, two pathobiological processes are pertinent. One, hepatocyte proliferation itself contributes to reduction in load of cccDNA within infected hepatocytes. In patients with advanced fibrosis or cirrhosis, the hepatocyte replicative senescence adds to the burden of cccDNA formation. Destruction of infected hepatocytes in the presence of potent replication suppression will help reduce cccDNA formation - a therapeutic approach that would require combination of multi-targeted treatment strategies. The cccDNA removal also occurs via non-cytolytic clearance of infected hepatocytes in the presence of antiviral cytokines, specifically interferon-α. It was shown that higher levels of interferon-α were associated with improved cccDNA clearance through triggering of non-cytolytic degradation of cccDNA from infected hepatocytes through induction of the nuclear deaminase A3A or A3B. However, such high doses in a clinical scenario can lead to adverse events and hence are impractical. Recently, the PASylation (addition of polypeptide comprising Proline, Alanine and Serine to increase plasma half-life) of interferon-α was found to improve antiviral effect without additional toxicity [ 55 - 59 ]. Figure Figure1 1 summarizes an updated schematic of the HBV life cycle and pertinent therapeutic targets.

HSPG - heparin sulfate proteoglycans, NTCP – Sodium taurocholate co-transporting polypeptide, LPS – lipopolysaccharide, UDCA - ursodeoxycholic acid, TUDCA – tauro-UDCA, GUDCA – glyco-UDCA, HBeAg – HBV envelope antigen, HBsAg – HBV surface antigen, ER – endoplasmic reticulum, RNA-H – ribonuclease H, rcDNA – relaxed circular DNA, cccDNA – covalently closed circular DNA, mRNA – messenger RNA, siRNA – small interfering RNA, pgRNA – pregenomic RNA, sgRNA – subgenomic RNA, CPAM - core protein allosteric modulator, Pol – polymerase, L – large HBsAg, S – small HBsAg, M – medium HBsAg
Updates on immunopathogenesis, genetic variants and applied biology
Host immune response against HBV infection includes innate immunity and adaptive immunity. The former includes downstream responses that are activated by pattern recognition receptor (PRR), natural killer (NK) cells, NK-T cells, and monocytes and macrophages; while the latter includes cluster of differentiation (CD)4 + T lymphocytes, CD8 + T lymphocytes, and B lymphocytes. In chronic HBV infection, the virus limits and evades antiviral effects of the innate immune and adaptive immune system through various mechanisms, resulting in continuous replication associated with dysfunction of various immune cells. HBeAg is immunomodulatory and is involved in antigen presentation and recognition by CD4+T cells. In HBeAg negative HBV infection (associated with pre-core stop codon mutation), there is rapid progression, cirrhosis and liver cancer development due to amelioration of host innate immune functions. Similarly, the HBV core promoter mutation in enhancer II which results in enhanced viral replication is accompanied by a reduction or loss of HBeAg leading to fulminant or progressive chronic hepatitis. The HBsAg mutant - defect in S region to arginine at amino acid position 145 and loss of group-specific antigenic determinant a (target of vaccine response) - escapes immune surveillance and infection even in the presence of antibodies to surface antigen and also development of occult HBsAg negative HBV infection. Vaccine-escape mutations occurred in particular when lamivudine (currently not utilized) was used in the long term [ 60 - 64 ]. The size of exposure or inoculum determines HBV persistence and clearance. Low-dose inoculum leads to a massive spread of the virus in all of the hepatocytes and viral persistence; whereas high-dose inoculum showed a limited spread of the virus to hepatocytes and rapid viral clearance. This phenomenon depends on the synchronized effector activity of CD4+ and CD8+ T cells. Exhaustion and depletion of CD4+ T cells in limited exposure infection along with synchronized influx of HBV-specific CD8+ T cells in the liver promotes viral persistence. In high viral load, interferons-α/β suppress viral replication through transcriptional and post-transcriptional modification. In early infection and low viral load, HBV utilizes host interferon response to promote viral persistence via stimulation of enhancer I in the genome which interacts with STAT3 and hepatocyte nuclear factor 3γ (HNF3γ). Innate immune activation functions through PRRs recognizing pathogen-associated molecular patterns (PAMPs). These include: a. Toll-like (TLRs; TLR2 activation promotes pro-inflammatory cascade for viral clearance, TLR4 activation through HBsAg related dendritic cell, soluble CD14 dependent cytotoxic T cell mechanism) receptors; b. Retinoic acid-inducible gene I (RIG)-like (dual antiviral effect on pre-genomic RNA through type III interferon induction and HBV polymerase interaction) receptor; c. Nucleotide-binding oligomerization domain-containing protein (NOD)-like, C-type lectin receptors and d. DNA-sensing (cytosolic cGAS recognize HBV DNA, suppress interferon suppressing regulatory factor 3, promote viral persistence) receptors.
Some preclinical studies have shown that, in early HBV infection, PRR-mediated innate immune responses are not activated - the stealth virus phenomenon where the virus interferes with innate signaling pathways to attenuate intrinsic antiviral immune responses [ 65 - 67 ]. HBsAg and HBeAg, in a dose-dependent manner, via interference with c-Jun N-terminal kinases (JNK) activation, inhibits expression of TLR2 mediated IL-12 and tumor necrosis factor-α (TNF-α) production in monocytes and macrophages. HBV also suppressed nuclear factor kappa B (NF-κB), extracellular signal-regulated kinase (ERK)1/2, blocked myeloid differentiation primary response 88 (MYD88) protein expression and inhibited type 1 interferon induction (via HBxAg protein). Recombinant HBx protein-based small interfering RNA (siRNA, short interfering RNA or silencing RNA) recovered interferon-1 activity by activating RIG-1 pathway. Nonetheless, detailed molecular determinants for potential recognition of HBV PAMPs by PRR still remain to be elucidated. This would increase the therapeutic armamentarium to include PRR agonists that would help in viral clearance [ 68 - 70 ].
Cellular Level Immune Activity in HBV Infection
NK cell dysfunction is also central to viral persistence in HBV infection. The ability of myeloid DCs to activate NK cells is impaired due to weak action in decreasing activating cytokines (IL-6, IL-12, IL-18) resulting in reduced secretion of interferon-γ and lowered activity of interferon-α. IL-10 secretion from Kupffer cells (liver resident macrophages) promotes cytokine blunting and hence lowers NK activation. IL-10 is an immune-suppressive cytokine (also secreted by virus-specific B lymphocytes) that maintains the immune tolerance during persistent HBV infection. Programmed death-ligand 1 (PD-L1) on suppressive monocytes also inhibit autologous NK cell activation. Reduction in the NK-cell mediated cytotoxic prowess and IFN-γ production contribute to HBV persistence. The expression of activating receptors on NK cells such as the NKG2D and 2B4 are also reduced in chronic HBV infection.
The myeloid-derived suppressor cell (MDSC) with predominant granulocytic subset (gMDSC) and monocytic MDSC (mMDCS) has an inverse relation with T cell function and hepatitis in chronic HBV infection. MDSCs potentiate CD4+ and CD8+ T cell responses via arginase-dependent pathways. High arginase levels reduce amount of arginine required for lymphocyte physiology and growth and resulting in lymphocyte dysfunction. Disruption of MDSC differentiation and T-regulatory cells (Tregs) resulted in immunosuppressive cytokine reduction which inhibited HBV replication. NKT cells of a special subset of T lymphocytes that express surface markers of T lymphocytes and NK cells - the invariant NKT cells (iNKT) lose functionality in the presence of HBV infection through an increase in T-cell immunoglobulin and mucin domain-3 (Tim-3) and programmed cell death protein 1 (PD-1) - antiviral treatment or Tim-3 blocking restores immune function of iNKT cells and improves viral clearance [ 71 - 74 ].
Interferon-γ secreted by lymphocytes in HBV infection induces Kupffer cells to produce chemokine (C-X-C motif) ligand 9 (CXCL9) and recruits HBV-specific CD4 + T lymphocytes to enter the liver for apoptosis, leading to chronic HBV. Defects in CD8 + T-lymphocyte functions through multiple pathways [blunted cytokine responses, T-lymphocyte depletion, high expression of co-inhibitory molecules such as Tim-3, PD-1 and CTLA-4, upregulation of TNF-related apoptosis-inducing ligand (TRAIL), arginase secretion] result in reduced HBV clearance from hepatocytes. PD-1 blockade can partially restore B cell function and CD 8+ T cell functions for viral clearance. Higher level of T helper cell 17 (Th17) lymphocytes (secretes IL-17, IL-21, IL-22) in the liver and peripheral blood was associated with acute and acute on chronic liver failure due to HBV. In chronic HBV infection, a follicular helper T (Tfh) cells response to HBsAg was required for HBV clearance which was blocked by Treg cells [ 75 - 82 ].
Endoplasmic reticulum (ER) stress also plays an important role in viral persistence. In HBV-infected cells, a large number of viral surface proteins are folded in ER during the replicative phase, resulting in disruption of ER homeostasis and ER stress. This is identified as ground-glass hepatocytes that accumulate ER mutant surface proteins (pre-s1 and pre-s2 mutants) which represent ER hypertrophy. ER stress leads to activation of ER degradation enhancers and hence reduction in the immune responses for viral clearance. The intracellular imbalance in favor of L-HBs compared with M- and S-HBs leads to ER stress, which can trigger cellular signals for apoptosis or uncontrolled cellular growth [ 83 ]. HBV RNA directly degrades host micro-RNA (miRNA, which is non-coding) leading to reduction in levels of miRNA-122 [block fibrosis by blocking collagen synthesis via transforming growth factor beta (TGF-β) pathway], miRNA-15 family and miRNA-let-7 family which lead to increased HBV replication, liver fibrosis and carcinogenesis [ 84 - 90 ]. Based on our current understanding of HBV immunopathogenesis, novel treatment strategies for enhancing chances for clinical cure of chronic HBV infection include PRR, TLR7 or RIG-I agonists (increases innate immune responses), PD-1 blockade (immune checkpoint blockers), therapeutic vaccines (based on miRNA), and chimeric antigen receptor T lymphocytes that improve adaptive immune responses for enhancing viral clearance. Figure Figure2 2 summarizes an updated schematic of the pertinent immunopathogenic processes and therapeutic targets in HBV infection.

HSPG - heparin sulfate proteoglycans, NTCP – Sodium taurocholate co-transporting polypeptide, TLR – toll-like receptors, CD – cluster of differentiation, PRR – pattern recognition receptors, PD - programmed cell death protein, CTLA - cytotoxic T-lymphocyte-associated protein 4, Tregs – T-regulatory cells, IL – interleukins, TIM-3 - T cell immunoglobulin and mucin domain-containing protein 3, miRNA – micro-RNA, NK – natural killer cells, MDSC - myeloid-derived suppressor cell, HBeAg – HBV envelope antigen, HBsAg – HBV surface antigen, ER – endoplasmic reticulum, RNA-H – ribonuclease H, rcDNA – relaxed circular DNA, cccDNA – covalently closed circular DNA, mRNA – messenger RNA, siRNA – small interfering RNA, pgRNA – pregenomic RNA, sgRNA – subgenomic RNA, CPAM - core protein allosteric modulator, Pol – polymerase, L – large HBsAg, S – small HBsAg, M – medium HBsAg
Updates on HBV genotypes and their clinical importance
Currently, 10 genotypes of HBV exist, with additional subtypes (mutations or recombinant strains), which are identified by the letters A to J and numbered respectively, which, through genetic mutations and the lack of proofreading in reverse transcriptase, have evolved over the long term, creating challenges to their elimination. An example is the HBV genotype B2 which is a recombinant, with majority of the genetic framework from HBV genotype B, and the precore/core region from genotype C. Coinfection with different HBV genotypes and intergenotypic recombination of HBV strains are extensively documented. Most commonly associated recombinants include genotypes B/C or A/D. Each genotype is classified by an 8% or more divergence in the nucleotide sequence of the genome. Genotypes A to D are the four predominant genotypes; B and C are most common in eastern and southeastern Asia, A and D are found in North America, Africa and Europe and genotype E is found in West Africa. Genotypes A and B have a greater response to interferon therapy than C and D, but none of the genotypes have differential responses to oral antivirals [ 91 - 93 ]. Delayed HBeAg seroconversion and a higher risk of reactivation in the HBeAg-negative phase were notable in HBV patients with genotype C who also have more advanced fibrosis. Liver cancer develops in young patients without cirrhosis who harbor HBV genotype B-related infection. Patients with HBV genotypes C and D, compared with genotypes A and B, have late or absent HBeAg seroconversion after multiple hepatitis flares that accelerate progression of liver disease, conferring worse clinical outcome. HBV genotypes are also associated with specific virological manifestations such as higher frequency of basal-core promoter A1762T/G1764A variants, pre-S deletion mutations, greater viral replicative burden, expression of intracellular HBV DNA and core protein expression and HBeAg secretion in genotype C when compared with other genotypes. In a systematic review and metanalysis, authors found that the blood group B was associated with a lower risk of HBV infection and persons with blood group O had a 12% increased risk of HBV infection in endemic regions [ 94 - 97 ].
Evaluation and treatment of HBV related liver disease
Current Approaches to Diagnosis and Evaluation
Presence of HBsAg indicates acute or chronic infection and is the first serologic marker to appear. HBV infection is considered chronic if HBsAg persists beyond six months. The HBeAg indicates active replication while its absence can also indicate mutations in the pre-core region of the e-antigen that prevent production of HBeAg. Antibody response to HBeAg (anti-HBe) indicates that the virus is non-replicative, but is also seen among HBV patients with HBeAg mutation with active disease. Antibody to HBc antigen can be present in acute infection and reactivation (IgM) and with past exposure to HBV (IgG). It can be seen in solitude when antibody response to HBsAg is waning. Patients who are HBsAg-positive and anti-HBc-positive need further evaluation for initiation of treatment. Those who are anti-HBs-positive and anti-HBc-positive are considered to have infection in the past and currently resolved. Nonetheless, in these patients infection may remain latent, only to reactivate under special circumstances (immunosuppression, spontaneous mutation) along with re-emergence of HBsAg. Patients who are HBsAg, anti-HBs, and anti-HBc negative do not possess immunity needs vaccination. Patients who are only anti-HBs-positive are immune or have undergone vaccination [ 98 - 100 ].
In patients who are HBsAg and antibody (total) to HBcAg positive, further differentiation is made on the presence or absence of HBeAg after which classification into chronic infection and chronic hepatitis is made for treatment decisions. Patients with chronic infection have normal alanine aminotransferase (ALT) and no or minimal liver damage (fibrosis grade <2) while those with chronic hepatitis have elevated HBV DNA and ALT with ongoing necro-inflammatory liver damage or significant fibrosis (grade ≥2), for whom treatments are to be directed [ 101 , 102 ]. The alanine transaminase concentrations generally correlate with hepatic necroinflammation in HBV patients. High-normal ALT levels ranging from 40 to 70 IU/liter are linked to cirrhosis and liver-related deaths. Currently, guidance on recommendations suggests that the ALT cutoffs should be 35 U/liter for males and 25 U/liter for females and significant elevation is considered two times the upper limit of normal (ULN). Even though percutaneous liver biopsy and histological interpretation is the gold standard for fibrosis assessment, its use in clinical practice is limited and follow-up biopsies are not routinely employed due to patient unacceptance. In this regard, assessment of hepatic fibrosis by non-invasive modalities is suggested. These include shear wave elastography (transient, acoustic radiation force impulse, or multidimensional) as well as magnetic resonance elastography (MRE). Among these the FibroScan® transient elastography is the best validated worldwide. In obese patients and those with ascites, MRE is to assess liver stiffness measurement (LSM) with phase contrast imaging is suggested, which can also stage even mild fibrosis, but it is less cost effective, not well tolerated and more time consuming than ultrasound methods. Other validated serum biomarker combinations for diagnosis of significant fibrosis in HBV patients include the aspartate aminotransferase (AST)-to-platelet ratio index (APRI, low sensitivity in African patients), the Forns index, Fibrotest®, Fibrosure™, Fibrometer® and enhanced liver fibrosis (ELF™) score [ 103 - 106 ].
There are three types of treatment end points or cure in HBV infection. In functional cure, there is resolution of clinical infection which is sustained off drug treatment - no inflammation, normal ALT level and normal liver biopsy, HBsAg seroclearance (equal to 0.05 IU/ml in serum) with or without the emergence of anti-HBs. Protective immunity is when the anti-HBs level is greater than 10 IU/ml. Complete cure is virologic cure, consisting of all the elements of functional cure plus loss of cccDNA within the liver. In clinical practice, most of the treated patients fall into an interim cure period in which there is disease inactivity - absence of inflammation (normal ALT level and liver biopsy), low or undetectable HBV DNA level in the presence of HBsAg positivity. In this situation, a patient with chronic hepatitis is effectively down staged to one with chronic infection. An HBsAg level of 100 IU/ml in Asian HBeAg-negative patients is predictive of spontaneous HBsAg seroclearance within six to eight years. Inactive - low replicative chronic HBV patients have high rates of spontaneous HBsAg seroclearance, - 8.1% and 44.7% after 10 and 25 years of follow-up, respectively [ 107 - 110 ]. In a nutshell, HBsAg patients who require treatment with antiviral agents include patients with chronic hepatitis, those with cirrhosis (any level ALT, detectable HBV DNA and decompensated patients irrespective of DNA and ALT levels), those with hepatocellular carcinoma (HCC), HIV coinfection, on chemotherapy or biologic and immunomodulatory agents, women in the third trimester of pregnancy if HBV DNA is greater than 200,000 IU/mL and those with extrahepatic manifestations such as glomerulonephritis and vasculitis [ 111 , 112 ]. A summary of various international guidelines for the management of chronic HBV is shown in Table Table1 1 .
WHO – World Health Organization, ATA – American Treatment Association, APASL Asia-Pacific Association for the Study of Liver, EASL – European Association for the Study of Liver, AASLD – American Association for the Study of Liver Diseases, APRI - aspartate transaminase (AST) to Platelet Ratio Index, HBV – hepatitis B virus, ALT – alanine aminotransferase, ULN – upper limit of normal, ETV – entecavir, TDF – tenofovir disoproxil, TAF – tenofovir alafenamide, IFN – interferon, LAM – lamivudine, ADV – adefovir, LdT – telbivudine
| Management | WHO 2015 | ATA 2015 | APASL 2015 | EASL 2017 | AASLD 2018 |
| When to initiate | Compensated or decompensated cirrhosis (or APRI > 2 in adults) Age >30 yr, persistently abnormal ALT, and HBV DNA > 20,000 IU/ml HBV DNA not available, then on bases of persistently abnormal ALT levels | ALT level >2 x ULN and HBV DNA >2,000 IU/ml Compensated/decompensated cirrhosis with detectable HBV DNA | ALT >2 x ULN and HBV DNA >2,000 IU/ml in HBeAg negative or > 2,000 IU/ml in HBeAg-positive Compensated or decompensated cirrhosis with detectable HBV DNA | HBV DNA >2,000 IU/ml, ALT >ULN and moderate liver necro-inflammation or fibrosis (F2 minimum) Compensated or decompensated cirrhosis with detectable HBV DNA HBV DNA >20,000 IU/ml and ALT ≥2 x ULN HBeAg positive, high HBV DNA level, and age 30 yr Family history of HCC, cirrhosis, extrahepatic manifestations | ALT >2 x ULN and HBV DNA >2,000 IU/ml in HBeAg negative or > 20,000 IU/ml in HBeAg positive Cirrhosis with HBV DNA > 2,000 IU/ml Age 40 yr, family history of HCC, previous treatment, extrahepatic disease |
| What to treat with | ETV, TDF | ETV, TDF, Peg IFN-α | ETV, TDF, Peg IFN-α or LAM ADV and LdT (less preferred) | ETV, TDF, TAF, Peg IFN-α | ETV, TDF, Peg IFN-α |
| When to stop | Lifelong treatment in cirrhosis Stop treatment if non-cirrhotic, HBeAg seroconversion, or persistently normal ALT levels with or without undetectable HBV DNA Stop treatment in case of persistent HBsAg loss with 1 yr of consolidation therapy | HBsAg loss for 6–12 mo | HBsAg loss for at least 12 mo Non-cirrhotic HBeAg seroconversion and undetectable HBV DNA after minimum 1 yr (preferably 3 yr) of consolidation therapy Non-cirrhotic HBeAg negative and undetectable HBV DNA for ≥2 yr | HBsAg loss Non-cirrhotic HBeAg positive with seroconversion and undetectable HBV DNA after 12 mo of consolidation therapy Noncirrhotic HBeAg negative with undetectable HBV DNA for 3 yr | HBsAg loss Lifelong in cirrhosis |
| Retreatment | Reactivation of HBV | Relapse of HBV with respect to HBV DNA and ALT levels (specific levels not provided) | None | Similar to treatment-naive patients | None |
It is important to note that among HBsAg-positive cases who additionally suffer from obesity or metabolic syndrome, the risk of development of cirrhosis is higher than those with HBV infection alone [ 111 , 112 ]. In these patients, if HBV DNA level is low or undetectable then the abnormal ALT level may be due to non-alcoholic fatty liver disease (NAFLD) which requires targeted treatments and lifestyle modifications with close follow up. However, in those with increasing ALT, only a liver biopsy can help differentiate between NAFLD-related liver disease or HBV-associated necro-inflammation. Current literature suggests that patients in the immune tolerance phase or HBeAg positive chronic infection (very high HBV DNA >10 7 IU/mL and normal ALT) if aged above 30-40 years benefit from antiviral therapy irrespective of other standard inclusions for treatment initiation. Nonetheless, one must be aware that spontaneous HBeAg and HBsAg clearance with remission of liver disease can occur in 70 - 80% of patients at median follow up of approximately 10 to 20 years [ 113 - 115 ].
New updates on diagnosis and monitoring
The HBV core-related antigen (HBcrAg) is a new indicator that encompasses amino acid sequence common to HBeAg and HBcAg as well as the 22-kDa precore protein. HBcrAg positivity correlates with intrahepatic HBV DNA and pregenomic RNA levels among patients on antiviral treatment. This makes HBcrAg measurement a good serum marker of the active transcriptional activity of liver cccDNA and higher levels correlate with increased risk of hepatocellular carcinoma [ 116 - 118 ]. Another potentially new viral marker for future clinical use is measurement of HBV RNA which has been shown to provide significant insights into antiviral treatment response and cessation decision; identification of functional cure in chronic HBV infection; risk for HBV-related liver cancer and levels of intrahepatic HBV cccDNA. Particularly, the HBV pre-genomic RNA reflects viral replication activity and could be a very valuable tool for monitoring the effect in patients receiving novel anti-HBV therapies. These viral molecules are promising as surrogate markers for HBV viral activity, and when used alongside standard biomarkers, they allow for better assessment of HBV infection and treatment responses [ 119 - 121 ]. Since carcinogenesis is an important aspect in the natural history of HBV-related infection, the novel HBV DNA quantitation-time index (HDQTI), comprising HBV DNA quantitation and follow up, was found to predict HBV associated liver cancer prognosis identified a cut-off value at 34. The HDQTI also predicted cancer recurrence and the need for shorter surveillance intervals with appropriate imaging in patients with a high score [ 122 ]. Liver biopsy can be avoided in a significant number of patients with use of the combined ELF™ (based on Fibroscan®) algorithm. The optimal cut‐off values of ELF™ were 8.4 to exclude advanced fibrosis, and 10.8 to confirm advanced fibrosis and LSM ≤6.0 kPa and ≤7.5 kPa excludes ≥F3 fibrosis while LSM>9.0 kPa and >12.0 kPa diagnose ≥F3 fibrosis in normal and elevated (1-5× ULN) ALT, respectively [ 123 ].
Streamlining the HBV diagnostic process to identify those who would benefit from screening, surveillance or therapy through artificial intelligence-related machine learning (ML) is a novel technique. With the help of ML, a predictive model for inflammation grades of chronic HBV was proposed utilizing a combination of gene expression data and three clinical parameters (ALT, AST, HBV DNA) over which a user-friendly web tool (LiveBoost™) was applied for the clinical prediction of hepatic fibrosis. It was demonstrated that the ML system outperformed FIB-4 scoring in predicting advanced hepatic fibrosis. Additionally, an artificial neural network (ANN) model was found to be effective in diagnosing liver fibrosis regression in HBV patients on therapy. ML-based models were also found to accurately identify persons at risk for HBsAg positivity, predict HBsAg seroclearance, predict treatment decisions in HBV carriers; 28- and 90-day mortality of HBV related ACLF, determine viral variants; and interactions between viral and host proteins to map pathways in hepatocellular carcinoma [ 124 - 128 ].
Current measures and updates on prevention and treatment
The immunogenic first-generation active HBV vaccines were made from materials extracted directly from plasma of chronically HBV-infected patients. It was not the virions that were utilized, but the large amounts of non-infectious spherical viral particles in carrier plasma, which were easily separated by biophysical methods. After cloning the HBV genome, large-scale production of spherical viral particles within recombinant yeast cells (yeast-derived second-generation vaccines) showed comparable protection with plasma-derived vaccines. Vaccine response rates with yeast-derived vaccines are>99% among infants and adolescents, but insufficient in 5% of healthy adults. The majority of yeast-derived vaccines consist only of the S-HBs of the globally underrepresented HBV genotype A2, dominant only in Northern Europe and North America. Very high anti-HBs titers (> 1,000 IU/L) are protective, but low or waning anti-HBs-titers over time increase risk of breakthrough infection with antigenically distant HBV genotypes (HBV genotypes B, C, D and F and at anti-HBs titers of less than 100 IU/L). The minimal infectious dose of HBV is as low as 16 virions (or 3 IU) when transmission occurs through HBV-contaminated blood transfusions [ 129 - 132 ].
Current therapies for the management of HBV include interferon-α (standard or pegylated) and orally administered NAs. First-line therapy should be with an oral antiviral with a strong genetic barrier to viral resistance such as either entecavir, tenofovir disoproxil (TDF) or tenofovir alfenamide (TAF, prodrug of TDF with more stable concentration in serum hence lower dose and less systemic exposure). Short-term treatment with NA is feasible in HBeAg-positive patients experiencing seroconversion to anti-HBe during treatment. A randomized controlled study (FINITE) analysed outcome when TDF therapy was withdrawn in a set of HBeAg negative patients who had achieved suppression of HBV DNA. Interestingly, 43% of patients achieved either HBsAg loss or suppressed DNA without any significant safety concerns [ 133 - 136 ].
Peginterferon for chronic HBV-related hepatitis is not widely used, even though the treatment period is finite (48 wks therapy). For HBeAg positive patients with low HBV DNA (<2 × 10 8 IU/mL), genotype A, and elevated serum ALT (> 2-5 times ULN) along with necroinflammation on liver biopsy, peginterferon-α could be used as first-line antiviral agent. HBeAg negative, genotype D patients who do not experience decrease in HBsAg levels and 2 log10 IU/ml reduction of HBV DNA at 12 weeks peginterferon-α treatment are considered non-responders. The HBsAg level is useful for prediction and motoring of response to therapy with peginterferon. HBeAg-positive patients, with HBsAg level of 20,000 IU/ml at week 24 are considered non-responders and treatment can be stopped early. Peginterferon leads to higher rates of HBeAg and HBsAg loss at one year mainly in patients with genotype A infection. Overall rates of sustained response (HBeAg seroconversion and undetectable HBV DNA in HBeAg positive patients and DNA <2000 IU/mL in HBeAg negative patients) after a one-year course of treatment is 27-36% and 28% respectively. Combination of NA and peginterferon can be performed via two protocols - de novo combination or the simultaneous administration of the two agents in treatment-naïve HBV patients; and the sequential combination, which features “add-on” or “switch-to” strategy in those who are already on treatment with either drug. This strategy improves HBsAg loss. Nonetheless, the benefits are mainly limited to specific group of patients - those with low baseline HBsAg level and on-treatment HBsAg response, high baseline ALT and viral load and genotype A. Peginterferon should not be used in decompensated cirrhosis, but can be used with caution in patients with compensated cirrhosis [ 137 - 139 ].
For those with chronic HBV hepatitis and multiple drug-resistant virus strains, combination of TDF and entecavir seems to be an effective and safe rescue option. In general, after HBeAg seroconversion, the treatment should continue for at least one year and possibly an additional three years to achieve long-lasting response once therapy is discontinued [ 137 , 139 ]. This three-year continuation phase lowers relapse rates to <30% and hastens loss of HBsAg. Nonetheless, higher relapse rates after NA discontinuation occur in older patients and those with HBV genotype C infection. Ideally, NAs can be withdrawn in HBeAg negative patients only after confirmed loss of HBsAg, with or without antibody development. Recommendations from European and Asian countries suggest stopping of NAs in HBeAg-negative patients who have undetectable HBV DNA at three different times points, six months apart [ 139 , 140 ]. One should not stop NA in patients with cirrhosis. Long-term NA therapy can decrease the cccDNA pool of infected hepatocytes through inhibition of nucleocapsid recycling but cannot prevent the initial cccDNA formation in newly infected hepatocytes [ 140 , 141 ].
In patients with liver failure, benefits were observed in those with model for end-stage liver disease (MELD) score between 20-30, while the mortality rate in those with MELD >30 was >90% even in the presence of early antiviral treatment - these patients need early referral for liver transplantation [ 141 , 142 ]. After liver transplantation, antiviral therapy is indefinite, regardless of HBsAg, HBeAg, or HBV DNA status. For patients on immunosuppressive therapy, antiviral therapy should be continued for at least six to 12 and 12 to 18 months after completion of therapy, as per American and European guidelines respectively; longer duration specifically in those receiving rituximab [ 142 , 143 ]. For pregnant HBV patients, antiviral therapy should commence at 28 weeks gestation and continued 12 weeks post-partum. For patients with HCV and HBV co-infection, entecavir has the least drug-drug interaction and treatment can be started simultaneously. In people living with HIV and HBV co-infection, the treatment should include either TDF or TAF + lamivudine or emtricitabine along with other HIV drugs [ 142 - 144 ].
Future directions for HBV treatment
HBV entry inhibitors targeting NTCP receptors include myrcludex-B (also called bulevirtide, subcutaneous route) and cyclosporine A (CsA). The former, a synthetic lipopeptide derived from pre-S1 domain blocks infection of new hepatocytes and hinders amplification of intrahepatic cccDNA of infected hepatocytes. The latter, a cyclic non-ribosomal peptide inhibit NTCP transporter activity blocking viral entry into hepatocytes - but can impair sodium dependent bile acid uptake resulting in various adverse events. Nonetheless, recently discovered SCY450 and SCY995 derivatives of CsA do not impair bile acid uptake [ 145 , 146 ].
APOBEC3 cytidine deaminase activators (through lymphotoxin-β receptor, LTBR pathway) via engineered non-lytic T cells with HBV-specific T-cell receptors inhibited HBV replication in small animal models. LTBR agonists were found to degrade cccDNA and exhaust intrahepatic pool. Genome-editing using transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced short palindromic repeats/Cas9 (CRISPR/Cas9) or locked nucleic acid technology (LNA) can be used to target specific DNA sequences for cleaving. TALENs comprise a nonspecific Fok1 nuclease domain fused to a customizable (can be engineered to target and disrupt any specific DNA sequence) sequence-specific DNA-binding domain. However, the safety of such DNA sequence cleavage on ‘HBV integrated host genome’ and its consequences remain to be studied. HBV-specific CRISPR/Cas system-mediated removal of the full-length integrated HBV DNA and the disruption of HBV cccDNA in a stable HBV cell line was demonstrated recently. CRISPR/Cas9 system from Streptococcus pyogenes and S. thermophilus targeting conserved regions of the HBV genome resulted in degradation of > 90% HBV cccDNA by six days. Nonetheless, even though deep sequencing revealed that Streptococcus-CRISPR/Cas9 had no effect on the host genome, it induced intrinsic off-target adverse effects such as mutagenesis [ 147 - 150 ].
RNA interference (RNAi) by which siRNA produces gene silencing at the post-transcriptional level to downregulate the expression of targeted genes is another novel therapeutic area. The siRNA therapeutic, ARC-520 that targets cccDNA-derived pre-genomic RNA was found to reduce HBsAg levels in HBeAg-positive patients, but not HBeAg-negative patients as in the latter, HBsAg arises not from cccDNA pool, but from HBV DNA integrated with the host genome. The novel ARO-HBV (JNJ-3839), targeting two sources of HBsAg, pre-genomic and integrated DNA, is currently under evaluation. Another siRNA molecule called AB-729 using novel conjugated N-acetyl galactosamine delivery technology with strong anti-HBV activity which acts on all HBV RNA transcriptions is under evaluation [ 151 - 153 ].
Virus nucleocapsid assembly inhibitors/modulators (heteroaryldihydropyrimidines that function to misdirect formation of aberrant or non-capsid structures; and phenylpropenamides or sulfamoylbenzamides that function to produce dysfunctional intact empty capsids) limit HBV replication by causing capsid destabilization is under multiple trials. A novel acyclic nucleotide phosphonate called besifovir is recently approved for trial studies. The main adverse event noted was L-carnitine depletion (myonecrosis, hypoglycemia) in treated patients requiring supplementation. Another new lipid conjugated nucleoside analogue under clinical development is tenofovir exalidex (TLX) which shows enhanced hepatic targeting that maximizes liver activity while reducing systemic drug exposure and was found to enhance HBsAg loss and reduce the cccDNA amount. Another novel nucleoside analogue under phase II clinical study is CMX157 [ 154 - 157 ]. Inhibitors of ribonuclease H (α-hydroxytropolones, N-hydroxyisoquinolinediones and N-hydroxypyridinediones) limit degradation of HBV pre-genomic RNA during DNA minus strand synthesis thereby permitting plus-strand synthesis are a novel class of antiviral agents that block release of infectious virions and amplification/replenishment of cccDNA pool. Inhibiting HBsAg release ameliorates T cell tolerance, reduces T cell exhaustion and restores HBV-specific T cell-mediated immune response. Phosphorothioated oligonucleotide assembly blockers are nucleic acid polymers that prevent assembly of subviral particles which are the primary source of circulating HBsAg. Designated REP 301 and REP 401, these drugs used along with NAs or peginterferon may have better chances at promoting functional cure [ 158 - 162 ].
Based on our advances in understanding immunopathogenesis of HBV, multiple immune modulating therapeutic agents are under development which would help promote functional cure of HBV. TLR agonists (TLR-7 - oral vesatolimod or selgantolimod and TLR-8) induce endogenous interferon production, activate innate responses, leading to induction of interferon-stimulated genes (also called STING agonists) and other signaling cascades that inhibit HBV replication. Nonetheless, phase II studies have shown that even though T cell increase, NK cell responses and interferon signaling were improved with TLR agonists, reduction in HBsAg levels were not identified. This means that monotherapy with these agents is probably of low clinical relevance and hence combination strategies are warranted. Pattern recognition receptor agonists such as RIG-I and NOD-2 agonists activate interferon signaling pathways and proinflammatory cytokines that improve viral clearance. The RIG-I agonist, inarigivir soproxil, a novel oral modulator of innate immunity when used along with TDF significantly increased reduction of HBV replication, HBV RNA and HBsAg levels in a dose-dependent manner in both HBeAg-positive and HBeAg-negative patients [ 163 - 165 ].
PD-1, a highly expressed inhibitory receptor on HBV-specific T cells, along with increased expression of PD-L1 (PD-1 ligand), contributes to T cell exhaustion and high HBV replication in chronic HBV. Thus, PD-1/PD-L1 pathway blockers induce proliferation of HBV-specific T cells, thus restoring functioning T cells and helping control HBV. A pilot study showed that the PD-1 blocker nivolumab along with HBV therapeutic vaccine GS-4774 achieved significant and sustained HBsAg loss [ 166 - 168 ]. Finally, the novel therapeutic protein-based vaccines that include subunit vaccines (HBsAg+HBcAg called HeberNasvac) and antigen-antibody complex vaccines (HBsAg+HBV immunoglobulin) did not demonstrate favorable results due to non-induction of cytotoxic T cell responses. DNA-based vaccines encoding HBV envelope proteins such as INO-1800 (multi-antigen vaccine encoding HBsAg and consensus HBcAg sequence) that induce HBV-specific T cells; INO-9112 (encoding human IL-12); HB-110 (encoding HBsAg, pre-S1 Ag, HBcAg, HBV polymerase, human IL-12) are under evaluation. New vector-based vaccine GS-4774, a recombinant, heat-killed, Saccharomyces cerevisiae yeast-based vaccine expressing HBsAg, HBcAg, and HBx, did not provide significant reductions in serum HBsAg levels when used alone, but induced strong immunomodulatory effects when used along with TDF. The non-replicative adenovirus 5 vector vaccine TG1050 encodes a large fusion protein made of modified HBV core, HBV polymerase, and selected envelope protein domains. TG1050 was found to have a good safety profile and induced appreciable HBV-specific cellular immune response in early trials [ 169 - 171 ].
In another study, 12 chemical compound candidates for alpha-glucosidase inhibitors were identified from a library of chemical compounds and used to treat fresh human hepatocytes infected with HBV and monitored for their anti-viral effects. It was found that HBV replication was inhibited by one candidate, a tetramethylpiperidinol derivative in a dose-dependent manner, through interaction with HBV nuclear transcription factor Sp1 which was also associated with significant reduction of cccDNA production, compared to entecavir [ 172 ]. To summarize (Table (Table2), 2 ), novel therapeutic agents targeting functional cure for chronic HBV infection include entry inhibitors, cccDNA disruptors, translation inhibitors, capsid assembly blockers, polymerase and secretion inhibitors and state-of-the-art therapeutic vaccines [ 173 - 176 ].
HBV – hepatitis B virus, ccc- covalently closed circular, Ca – calcium, Mg – magnesium, TALENs - transcription activator-like effector nucleases, CRISPR-Cas9 - CRISPR-associated protein 9, TLRs – Toll-like receptors, PD – programmed death cell receptor, MVA - Modified Vaccinia Ankara, E – intramuscular – electroporation and intramuscular
| Type of drug | Name of drug | Clinical trial phase | Route | Mode of action |
| Entry inhibitors | Myrcludex-B / Bulevirtide | II | Subcutaneous | HBV entry blockade |
| Oligonucleotides | INOIS-HBVRx (GSK3228836), INOIS-HBVLRx (GSK33389404) | II, Preclinical | Subcutaneous | Antisense Antisense |
| Core protein allosteric modulators (CpAMs) | RO7049389 JNJ, 56136379 JNJ, 64530440 AB-506, ABI-H2158, ABI-H0731 (Vebicorvir), GLS4JHS, NVR 3–778 QL-007 | I to II | All oral | Core protein binding Assembly modulation Assembly modulation Core protein binding Core protein binding Core protein binding Core protein binding Assembly modulation Assembly modulation |
| HBx protein inhibitors | Nitazoxanide, CRV-431 | II, I | Oral Oral | cccDNA transcription Cyclophilin inhibitor |
| RNA interference | GSK3389404, ARO-HBV (JNJ-3989), AB-729, ALN-HBV (VIR-2218), ARC-520, DCR-HBVS | II to II | Subcutaneous or Intravenous Subcutaneous | RNA degradation RNA interference RNA interference RNA degradation RNA interference RNA interference |
| HBsAg release inhibitors | Nucleic acid polymers REP 2139 - Ca, REP 2165 - Mg | II | Intravenous Intravenous | Binding and prevents release of HBsAg surface protein |
| HBsAg neutralization | GC 1102 (Lenvervimab) | II | Intravenous | Neutralization and inhibiting reentry |
| Inhibitors of cccDNA | TALENs CRISPR-Cas9 | Preclinical | Unknown | cccDNA disruption cccDNA disruption |
| Cell intrinsic and innate immune responses (Toll-like receptor agonists) | RO7020531 Vesatolimod, GS-9620 Selgantolimod, GS-9688 AIC649 | I to II | Oral, Oral, Oral | TLR-7 agonist TLR-7 agonist TLR-8 agonist TLR-9 agonist |
| Immune checkpoint inhibitors | Nivolumab , Cemiplimab | I, I/II | Intravenous | PD-1 blockade PD-1 blockade |
| Therapeutic vaccines | TG1050/T101, INO-1800, ChAdOx1 HBV, Hep-Tcell, JNJ-64300535, GS-4774 | I to II | Subcutaneous or intramuscular | HBV proteins DNA plasmid Adjuvanted ChAd+MVA vector HBV peptide+ TLR9 adjuvant IC31 Electroporation DNA vaccine DNA vaccine |
Updates on HBV-related ACLF
Acute on chronic liver failure is a recently described entity in the natural history of cirrhosis, defined by acute insult leading to rapid hepatic decompensation, multiple organ failure and a high risk of short-term mortality, usually less than four weeks. Acute alcoholic hepatitis, drug-induced liver injury, infections and surgical stress are the most frequent precipitants for ACLF. Central to the pathophysiology of ACLF is the state of unchecked persistent inflammation and immune dysfunction with increased propensity to sepsis and organ failure. Reactivation of chronic HBV infection is an important and modifiable cause for ACLF [ 177 , 178 ]. Studies have reported an approximately 35% incidence of ACLF in patients with underlying HBV-related cirrhosis who suffered from acute decompensation. A large Chinese study estimated that the overall ACLF incidence rate over a 10-year period was 2.53 per 100,000 of the general population per year. The short-term mortality of HBV-associated ACLF is high, with 28-day mortality ranging from 40% to 50% depending on the diagnostic criteria as well as class and grade of ACLF. HBV infection was the most common acute insult precipitating HBV-associated ACLF in close to 60% of cases according to published data. The Asia-Pacific Association for Study of Liver ACLF Research Consortium (AARC) reported that acute viral hepatitis A and E contribute to 12.6% of acute insults, whereas a more recent study from the same group in ACLF revealed that complementary and alternative herbal medicines were the commonest cause for drug-induced ACLF [ 179 , 180 ].
In HBV-related ACLF, viral factors were found to have strong association with the development of the catastrophic syndrome. The HBV basal core promoter/precore mutations such as T1753V, A1762T, G1764A, A1846T, G1896A, and G899A correlated with an increased risk of HBV-related ACLF which was supported by the fact that ACLF patients had distinct quasi-species characteristics and higher complexities and diversification within the precore/core gene [ 181 ].
A genome-wide association study identified HLA-DR and rs3129859*C allele as the major locus for susceptibility to HBV-related ACLF. This allele was associated with prolonged prothrombin time, faster progression to ascites development and higher 28-day mortality in HBV-ACLF. The authors concluded that the HLA class II restricted CD4+ T-cell pathway on the immunopathogenesis of HBV-related ACLF [ 182 ]. Some studies have also shown that HBV genotype B was more susceptible to developing ACLF while this has been refuted in a large metanalysis [ 183 ].
Prognosis of HBV-ACLF can be ascertained by a variety of scoring systems which include the standard MELD and MELD-sodium (MELD-Na) scores, EASL chronic liver failure (CLIF)-Consortium-ACLF score (CLIF-C ACLF, better prognostic tool than MELD), integrated MELD, which includes hepatic encephalopathy and age, with an improved sensitivity of approximately 70-80% and the recently proposed AARC score, which integrates bilirubin, creatinine, prothrombin time, lactate, and hepatic encephalopathy which was found to be superior to the MELD in predicting outcomes [ 177 , 179 , 184 ]. Novel biomarkers such as serum M30 and M65 antigen and Golgi protein 3 - cell death markers were found to predict mortality in patients with HBV-ACLF. However, these have low sensitivity and are not routinely available for use [ 177 , 179 ]. Recently, multiple prediction models were devised by various authors looking at outcomes in patients with HBV-ACLF. One group found that hepatic encephalopathy, neutrophil percentage and platelet levels were independent risk factors for predicting the prognosis of HBV-ACLF. A new prediction model LR(p) was found to have better prediction accuracy than MELD, MELD-Na, and albumin-bilirubin (ALBI) scores [ 185 ]. In another study, multivariate analysis indicated that red cell distribution width, neutrophil-to-lymphocyte ratio (NLR), total bilirubin, serum creatinine and international normalized ratio (INR) were identified as risk factors for 90-day mortality in patients with HBV-ACLF. A risk assessment model, called the RNTIC, with cut-off value of 3.08 (sensitivity: 77.89%, specificity: 86.04%) was found to be more predictive of prognosis than MELD, MELD-sodium and Child-Pugh scores [ 186 ].
The NLR was also found to be an independent predictor of mortality in patients with HBV-ACLF undergoing treatment with artificial liver support systems (ALSS; combined plasma exchange and bilirubin adsorption performed with continuous renal replacement therapy machine and bilirubin absorbent column) suggesting that liver function in most patients with baseline NLR ≤3 recovered with ALSS treatment, and those with NLRs >6 require emergency liver transplantation [ 187 ]. The Chinese Group on the Study of Severe Hepatitis B (COSSH) found that regardless of the presence of cirrhosis, patients with HBV, total bilirubin ≥12 mg/dL and INR ≥1.5 should be diagnosed with ACLF. The COSSH prognostic score (0.741×INR+0.523×HBV-SOFA+0.026×age+0.003×TB) for short-term mortality was superior to five other scores based on both discovery and external validation studies [ 188 ]. Additionally, the HINT score, a novel prognostic score based on hepatic encephalopathy, INR, neutrophil count, and thyroid-stimulating hormone (TSH), was simpler and superior to the Child-Pugh, MELD, CLIF-SOFA, and CLIF-C ACLF scores and at least comparable with the COSSH-ACLF score. Sequential measurement of TSH was also helpful in prediction of poor outcomes in HBV-ACLF patients [ 189 ].
The age-bilirubin-international normalized ratio-creatinine (ABIC) score >9.44 was superior to the MELD score in predicting short-term survival (one and three month) in HBV-ACLF patients [ 190 ]. Another Chinese study found that the plasminogen (significantly lower in HBV-ACLF non-survivors than in survivors) was a good prognostic biomarker and sequential plasminogen measurements help identify clinical course of HBV-ACLF. A new score, known as “the P5”, incorporating plasminogen levels, hepatic encephalopathy occurrence, age, INR and total bilirubin, was significantly superior to the Child-Pugh, MELD and CLIF-C ACLF scores [ 191 ]. A study evaluating the ‘regenerating’ ability of the liver showed that overall survival rate within 180 days was 43.48%, and log10-AFP (alfafeto protein) ≥ 2.04 indicated a better prognosis with 76.9% specificity and 62.5% sensitivity for patients with HBV-related ACLF. A new prognostic model called the TACIA score (including total bilirubin, age, creatinine, INR and AFP) was found to predict short-term outcomes in patients with HBV-ACLF in that, patients with lower TACIA scores (<4.34) survived longer [ 192 ]. A Chinese group found that low AFP (log value <4.18) was associated with worse prognosis in patients with HBV-ACLF treated with liver support devices and a new model containing AFP, called ALSS‐prognosis model (APM - log value of AFP in microgram/L, INR, bilirubin, age, grade of encephalopathy and serum sodium), which showed potentially better prediction performance than MELD, MELD‐Na, and CLIF‐C ACLF score for short‐term outcomes [ 193 ]. A collaborative study on HBV-ACLF utilized the classification and regression tree (CART) analysis to group patients into low and high risk. CART analysis identified three factors prognostic of survival: hepatic encephalopathy, prothrombin time and total bilirubin level; and two distinct risk groups: low risk (28-day mortality, 10.2-39.5%) and high risk (63.8-91.1%). The CART model showed that patients lacking HE and with a prothrombin time ≤ 27.8 s and a bilirubin ≤5 mg/dl experienced less 28-day mortality after ALSS therapy. For HBV-ACLF patients with HE and a PT > 27.8 s, mortality was higher. The authors concluded that, for HBV-ACLF patients at high risk, unnecessary ALSS should be avoided [ 194 ]. The World Gastroenterology Organization (WGO) proposed classification according to the underlying liver disease: type A ACLF (patients with underlying non-cirrhotic chronic liver disease), type B ACLF (patients with previous compensated cirrhosis) and type C ACLF (patients with previous decompensated cirrhosis) was utilized to derive a new type-based prognostic model for HBV-related ACLF. Named the “model of ACLF prognosis based on type” or MAPT, the score, developed according to Cox proportional hazards multivariable analysis, included type of ACLF (A, B or C), age, total bilirubin, creatinine, INR and presence or absence of hepatic encephalopathy. The authors found MAPT to be superior to the CLF-C-ACLF, MELD and Child-Turcotte-Pugh scores in predicting 90-day mortality, with an area under the receiver operating characteristic curve of 0.802 with sensitivity of 71.77%, and specificity of 75.82% [ 195 ]. Recent high-quality studies have shown that TDF was superior to entecavir in HBV-ACLF (white blood cell count and HBV DNA reduction at two weeks independently predicted mortality at three months); ALSS treatment improved short-term survival and was associated with lower short-term death in patients with HBV-ACLF class 2; corticosteroid treatment did not improve transplant-free survival in patients with HBV-ACLF but, a metanalysis showed that it was effective in reducing jaundice, in-hospital mortality and ascites events; while a prospective multi-center clinical trial showed methylprednisolone therapy (1.5mg/kg/d day 1-3; 1 mg/kg/d day 4-5; and 0.5mg/kg/d day 6-7) increased six-month survival [ 196 - 200 ]. To summarize, specific mutations in HBV predispose to reactivation of the virus leading to ACLF in patients with HBV-related ACLF, which is also governed by HLA susceptibility and virus genotype in certain patient populations. Apart from the classical prognostic scores such as MELD and CLIF scores, newer prognostic tools like AARC, COSSH, ABIC, P5, MAPT and TACIA scores allow the clinician to identify patients who would benefit from early liver transplantation. Furthermore, the ALSS-prognosis model and the CART model help in identifying patients who would fail extracorporeal liver support therapy, in whom early liver transplantation is warranted. Improvement in decisions for clinical management, in the form of prediction and prognostic models and tools for assessing futility and early liver transplantation for HBV-ACLF, have become an important aspect other than the standard antiviral therapy regimen in this difficult to manage group of patients. Further clinically oriented studies and improved understanding of the virus biology and novel modifiable host factors will help the clinician in improving patient care for HBV-ACLF through an algorithmic approach that may become standard of care in the future.
Conclusions
Our understanding of the structure, biology, viral and immunopathogenesis in chronic HBV-related hepatitis has come a long way. Nonetheless, knowledge gaps still persist that currently limit our therapies toward a complete cure from this globally burdening disease. With the advent of new technologies and better tools such as next-generation sequencing, genome-wide association studies, single-cell RNA sequencing, gene editing and rigorous and well-coordinated collaborative clinical trials, we now understand viral and host-related factors in disease development and progression better than before. Novel modalities of treatments, such as viral RNA interference molecules, capsid assembly blockers, immune checkpoint inhibitors, HBsAg and cccDNA generation blocking molecules and innate immune system modulators, are in the pipeline and will eventually help us improve HBV-related patient outcomes.
The content published in Cureus is the result of clinical experience and/or research by independent individuals or organizations. Cureus is not responsible for the scientific accuracy or reliability of data or conclusions published herein. All content published within Cureus is intended only for educational, research and reference purposes. Additionally, articles published within Cureus should not be deemed a suitable substitute for the advice of a qualified health care professional. Do not disregard or avoid professional medical advice due to content published within Cureus.
The authors have declared that no competing interests exist.

IMAGES
VIDEO
COMMENTS
The core antibody to hepatitis B virus (anti-HBc) can be detected through immunoassays for total anti-HBc, which is able to detect both anti-HBc IgG and anti-HBc IgM. Anti-HBc IgM is the determinant of acute hepatitis B and is often the only sign that may be detected during the period of acute hepatitis B when HBsAg has become undetectable.
Abstract. Hepatitis B infection is still a global concern progressing as acute-chronic hepatitis, severe liver failure, and death. The infection is most widely transmitted from the infected mother to a child, with infected blood and body fluids. Pregnant women, adolescents, and all adults at high risk of chronic infection are recommended to be ...
Hepatitis B virus (HBV) infection is a major public health problem, with an estimated 296 million people chronically infected and 820 000 deaths worldwide in 2019. Diagnosis of HBV infection requires serological testing for HBsAg and for acute infection additional testing for IgM hepatitis B core antibody (IgM anti-HBc, for the window period when neither HBsAg nor anti-HBs is detected).
DOI: 10.1056/NEJMra2211764. VOL. 388 NO. 1. Chronic hepatitis B is caused by the hepatitis B virus (HBV), a hepatotropic DNA virus that can replicate at high levels and cause minimal disease or ...
Hepatitis B is inflammation of the liver in hominoidea, including humans, caused by the hepatitis B virus. The virus is transmitted by parenteral exposure, such as by exposure to infectious blood ...
Abstract. Hepatitis B virus (HBV) infects more than 300 million people worldwide and is a common cause of liver disease and liver cancer. HBV, a member of the Hepadnaviridae family, is a small DNA virus with unusual features similar to retroviruses. HBV replicates through an RNA intermediate and can integrate into the host genome.
In the United States, by CDC estimates, only one-third of HBV-infected patients or less are aware of their infection. Some reasons for these low rates of surveillance, diagnosis, and treatment include the asymptomatic nature of chronic hepatitis B until the very late stages, a lack of curative therapy with a finite treatment duration, a complex ...
Chronic hepatitis B (CHB) remains a major global health problem affecting more than 240 million people with high liver-related morbidity and mortality worldwide. This snapshot summarizes available HBV therapeutic strategies and highlights their corresponding stages of development from late preclinical to various clinical phases, which are indicated by different colors.
Kao JH, Chen PJ, Chen DS. 2010. Recent advances in the research of hepatitis B virus-related hepatocellular carcinoma: epidemiologic and molecular biological aspects. Adv Cancer Res 108:21 ... the Distinguished Research Paper Award for Young Investigators of the Hong Kong College of Physicians in 2010, 2013, 2014, and 2015; the Ten Outstanding ...
Chronic hepatitis B virus (HBV) infection affects about 296 million people worldwide and is the leading aetiology of cirrhosis and liver cancer globally. Major medical complications also include ...
Abstract. Hepatitis B virus (HBV) is a non-cytopathic, hepatotropic virus with the potential to cause a persistent infection, ultimately leading to cirrhosis and hepatocellular carcinoma. Over the ...
The prevalence of chronic HBV infection declined over time, particularly in children younger than 5 years, since the introduction of hepatitis B vaccination. HBV-related death rates also decreased, but HBV-related death counts increased as a result of population growth, ageing, and cohort effects. By 2019, many countries had met the interim seroprevalence target for children younger than 5 ...
Hepatitis B. Wen-Juei Jeng, George V Papatheodoridis, Anna S F Lok. Hepatitis B virus (HBV) infection is a major public health problem, with an estimated 296 million people chronically infected and 820 000 deaths worldwide in 2019. Diagnosis of HBV infection requires serological testing for HBsAg and for acute infection additional testing for ...
INTRODUCTION. Hepatitis B virus (HBV) has infected humans for at least the past 40000 years[] and is the 10 th leading global cause of death[].HBV is the only DNA-based hepatotropic virus that exerts many adverse effects on the infected cells leading to necroinflammation, fibrosis, and carcinogenesis[].The world health organization (WHO), in 2015 has estimated 257 million people infected with ...
Background: The hepatitis B virus (HBV) affects an estimated 290 million individuals worldwide and is responsible for approximately 900 000 deaths annually, mostly from complications of cirrhosis and hepatocellular carcinoma. Although current treatment is effective at preventing complications of chronic hepatitis B, it is not curative, and often must be administered long term.
Hepatitis B virus (HBV) infection remains a significant global health challenge, with chronic HBV leading to severe liver diseases, including cirrhosis and hepatocellular carcinoma. Current treatments often fail to eradicate the virus, highlighting the need for innovative therapeutic strategies. The CRISPR/Cas9 system has emerged as a dynamic tool for precise genome editing and presents a ...
The aim of this systematic review and meta-analysis is to evaluate available prevalence and viral sequencing data representing chronic hepatitis B (CHB) infection in Kenya. More than 20% of the global disease burden from CHB is in Africa, however there is minimal high quality seroprevalence data from individual countries and little viral sequencing data available to represent the continent.
Hepatitis B infections are a major cause of hepatocellular carcinoma (HCC), which kills more than 700,000 people globally every year. The virus drives cancer by rearranging human chromosomes ...
1. Introduction. Hepatitis B virus (HBV) infection remains a major health problem despite an extensive vaccination program worldwide. Globally, 260 million people are chronically infected with HBV and 890,000 are dying yearly from complications due to the advancement of HBV infection [1,2].HBV may play a role in the pathogenesis of chronic liver disease, cirrhosis, and hepatocellular carcinoma ...
Some 25% of carriers develop progressive liver disease. The annual mortality from hepatitis B infection and its sequelae is 1-2 million people worldwide.The following current topics are reviewed: immunization strategies against hepatitis B and the kinetics and antibody response; the controversy on screening blood donors for anti-core antibodies ...
1 INTRODUCTION. Hepatitis B virus (HBV) infection is one of the most common infectious diseases, impacting an estimated 4.1% of global population with approximately 316 million patients being chronic HBV carriers. 1 The prevalence of HBV-related diseases in the Asia-Pacific region is relatively high and can reach up to 15%, compared to that of America and Western European regions, where it is ...
Globally, 296 million people are infected with hepatitis B virus (HBV), and approximately one million people die annually from HBV-related causes, including liver cancer. Although there is a ...
Anti-HBe (antibody to e-antigen)-positive patients experience rapid progression to cirrhosis at an annual rate of 8 to 20%. Patients with cirrhosis progress to advanced liver disease and hepatic failure at a rate of 16% over five years [1 - 4]. Chronic HBV infection remains a huge burden on the patients, their family and the healthcare system ...
Hepatitis B virus infection is a major public health problem worldwide; roughly 30% of the world's population show serological evidence of current or past infection. Hepatitis B virus is a partly double-stranded DNA virus with several serological markers: HBsAg and anti-HBs, HBeAg and anti-HBe, and anti-HBc IgM and IgG. It is transmitted through contact with infected blood and semen.